
1 Quantum Si Investor & Analyst Day November 19, 2025

2 Investor Day Agenda 10:00–10:10 AMTackling the Complexity of the ProteomeJeff Hawkins, CEO 10:10–10:40 AM Proteus Program Update and Long-term Technology Roadmap Todd Rearick, CTO 10:40–11:05 AMThe Path to Detecting All 20 Amino AcidsJohn Vieceli, CPO 11:05–11:25 AMPost-translational Modification Analysis Solutions Brian Reed, Head of Research 11:25–11:45 AMThe Road to Proteus LaunchJeff Hawkins, CEO 11:45 AM–NoonQ&A SessionManagement

3 Proteins Are the Core of Biological Discoveries Across Many End Markets Academic Research Pharma + Biotech Industrial Defense Clinical Diagnostics Translational Research Agriculture
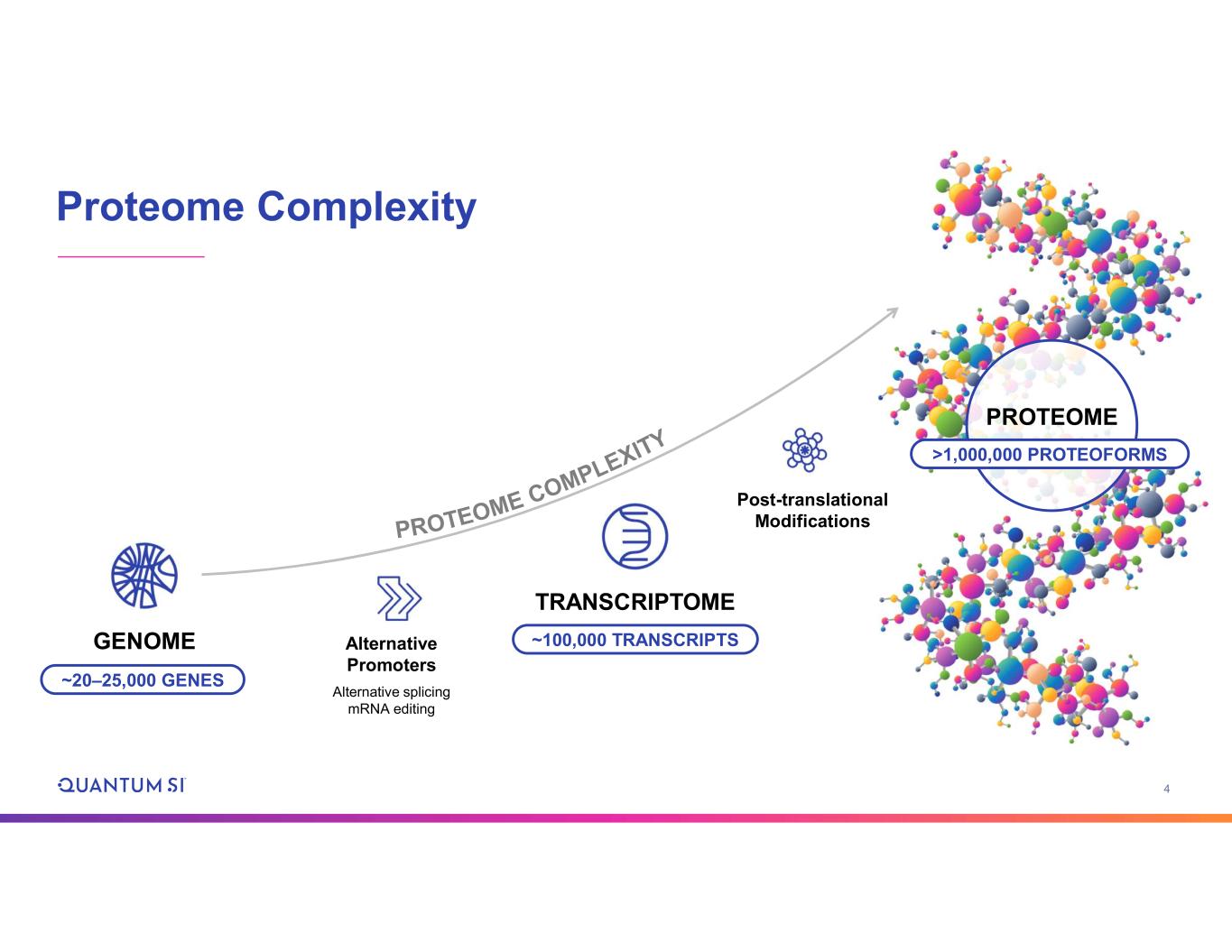
4 Proteome Complexity GENOME TRANSCRIPTOME Alternative Promoters Alternative splicing mRNA editing Post-translational Modifications ~20–25,000 GENES ~100,000 TRANSCRIPTS PROTEOME >1,000,000 PROTEOFORMS
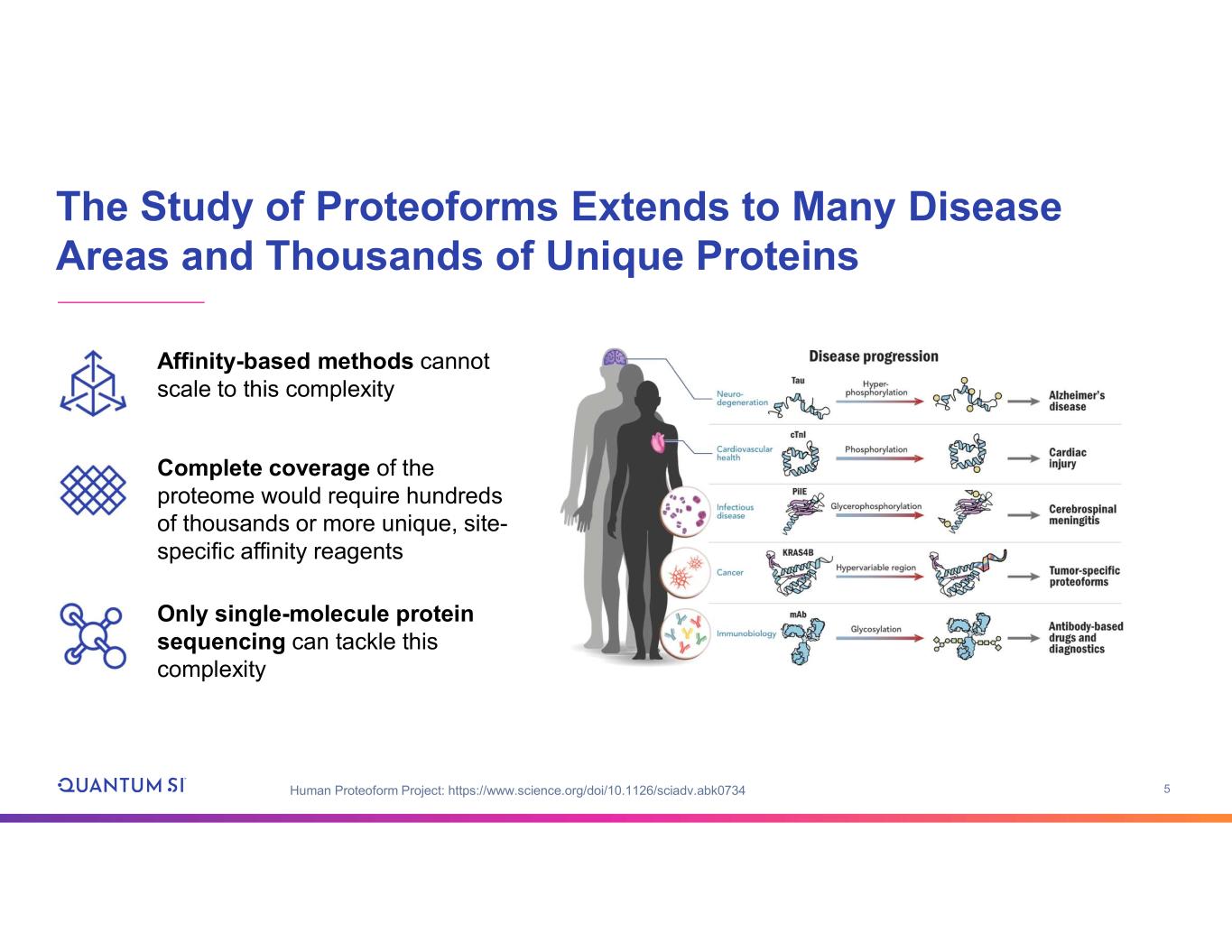
5 The Study of Proteoforms Extends to Many Disease Areas and Thousands of Unique Proteins Human Proteoform Project: https://www.science.org/doi/10.1126/sciadv.abk0734 Complete coverage of the proteome would require hundreds of thousands or more unique, site- specific affinity reagents Only single-molecule protein sequencing can tackle this complexity Affinity-based methods cannot scale to this complexity
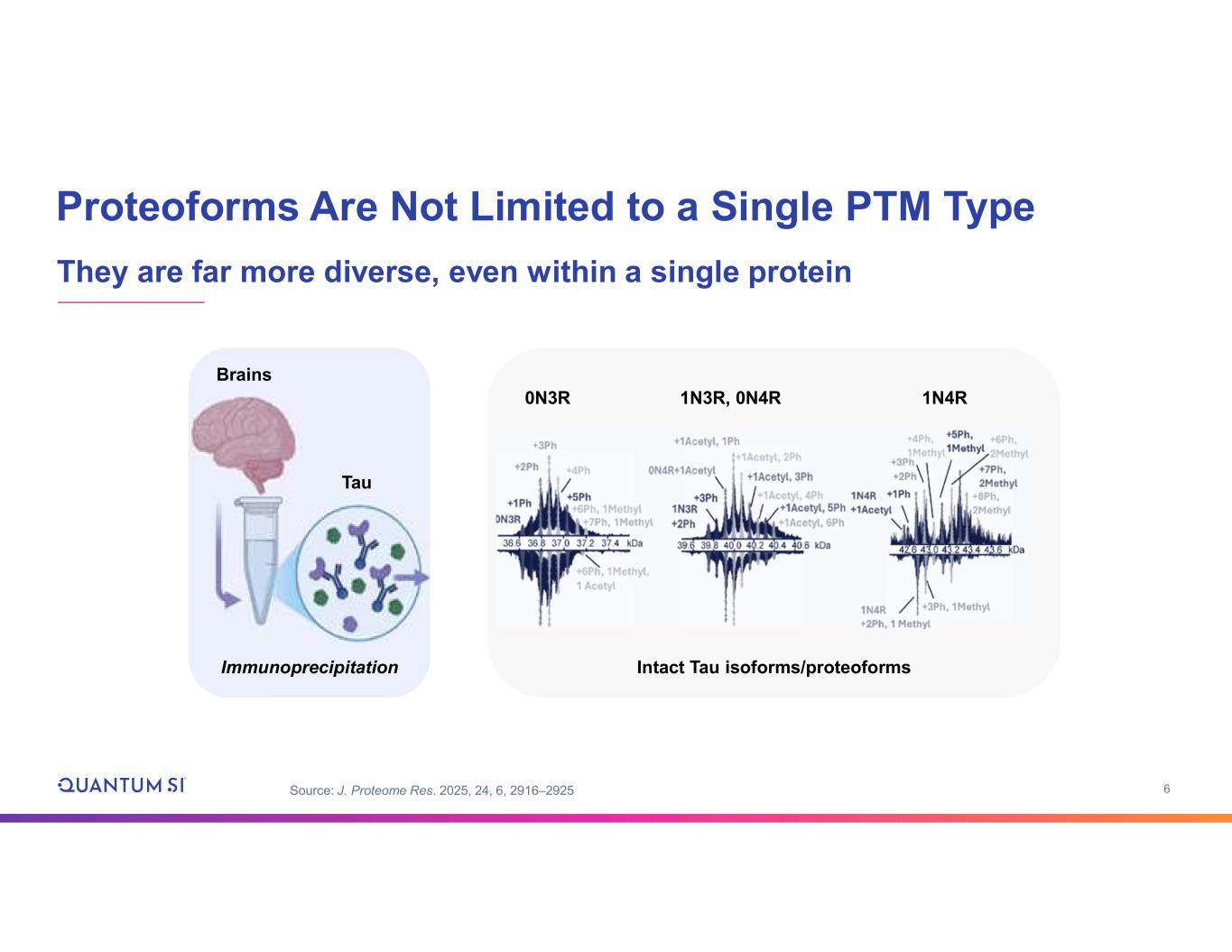
6 Proteoforms Are Not Limited to a Single PTM Type They are far more diverse, even within a single protein Source: J. Proteome Res. 2025, 24, 6, 2916–2925 Brains Tau Immunoprecipitation 0N3R 1N3R, 0N4R 1N4R Intact Tau isoforms/proteoforms
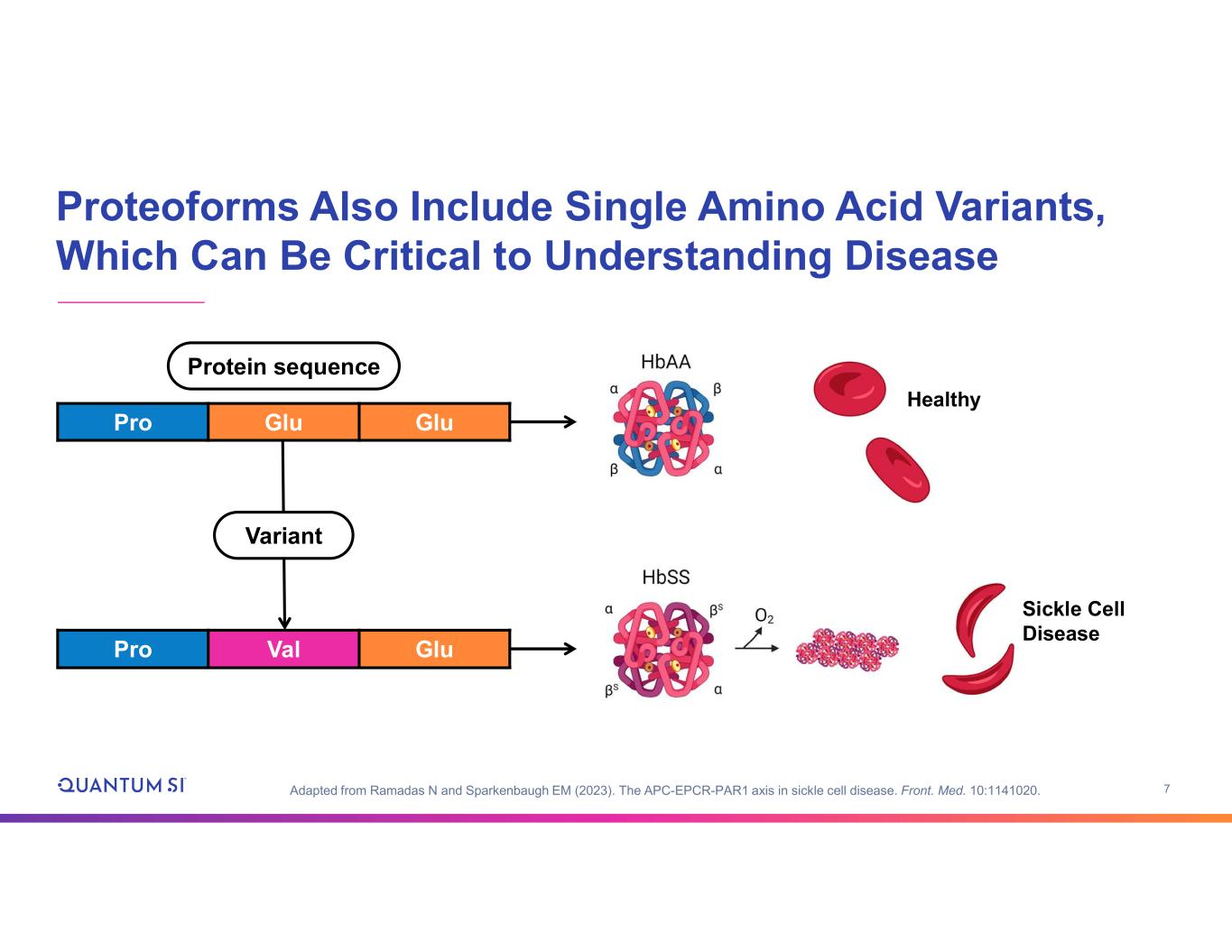
7 Proteoforms Also Include Single Amino Acid Variants, Which Can Be Critical to Understanding Disease GluGluPro GluValPro Healthy Sickle Cell Disease Variant Protein sequence Adapted from Ramadas N and Sparkenbaugh EM (2023). The APC-EPCR-PAR1 axis in sickle cell disease. Front. Med. 10:1141020.

8 Specialized Platforms With High Capital Costs Limit Adoption Outside of Core Labs in Proteomics Many specialized platforms needed to fully interrogate the proteome Technical tradeoffs when selecting between the breadth of protein coverage and depth of insights High capital costs with the top end instruments costs $1M or more each Manual laboratory and data analysis workflows limit the number of laboratories capable of performing proteomics

9 Moving Beyond the Core Lab: Proteus Will Enable Deep Protein Analysis and Accelerate the Field of Proteomics Automation simplifies laboratory workflow, increases throughput, and minimizes the need for specialized staff Affordable, allowing any lab — anywhere — to be a proteomics core lab Single-molecule protein sequencing will be the most versatile technology in proteomics — single AA variants, broad PTM coverage, and protein-agnostic

10 Themes of Today’s Presentations Proteus performance data and long-term technology roadmap The path to detecting all 20 amino acids A diverse set of tools to enable generalizable PTM analysis Road to Proteus launch and 2026 milestones

11 Proteus Program Update

12 Proteus Next-generation Architecture Platinum® Pro + 2M Chip • Integrated semiconductor consumable • Benchtop instrument with manual workflow Proteus + KinetIQ Array • Simple, passive consumable • Benchtop instrument with imaging system • Workflow automation

13 KinetIQ Array Simple, passive consumable • Low-cost fused silica die • 80M wells per device • Architecture scales to billions of wells Simple packaging • Inexpensive plastic assembly • Four flow cells, each with 20M wells • Features to support automation
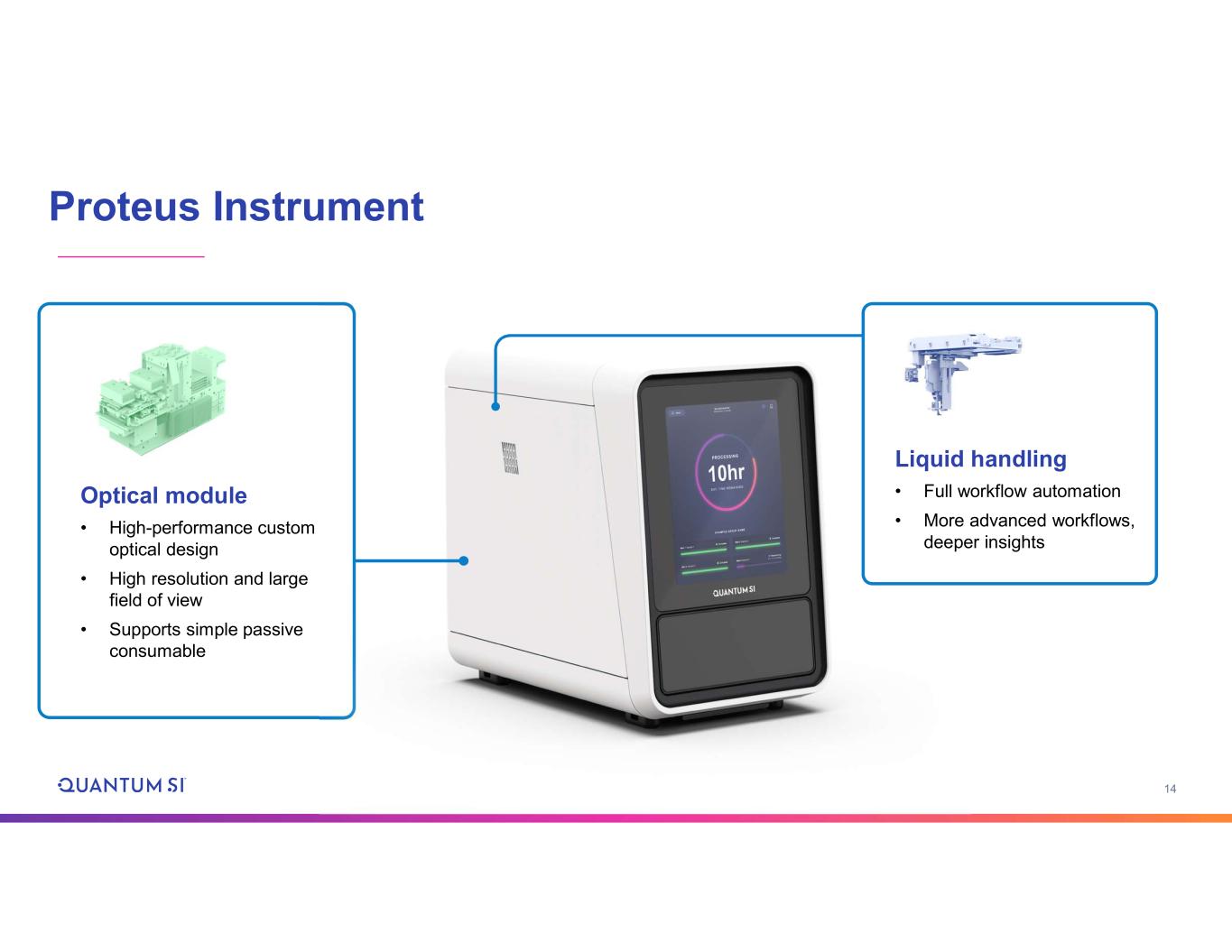
14 Proteus Instrument Optical module • High-performance custom optical design • High resolution and large field of view • Supports simple passive consumable Liquid handling • Full workflow automation • More advanced workflows, deeper insights
15 Proteus Program Status Key technical risks retired or substantially reduced Critical system components matured through several prototypes Integrated system design complete — building first systems now

16 Sequencing Workflow Automation is More Reproducible Than Current Manual Workflow Integrated System Liquid Handling • Same architecture as integrated product design • Entire sequencing workflow fully automated Prototype Liquid Handling Subsystem Data Automated vs manual workflow alignments
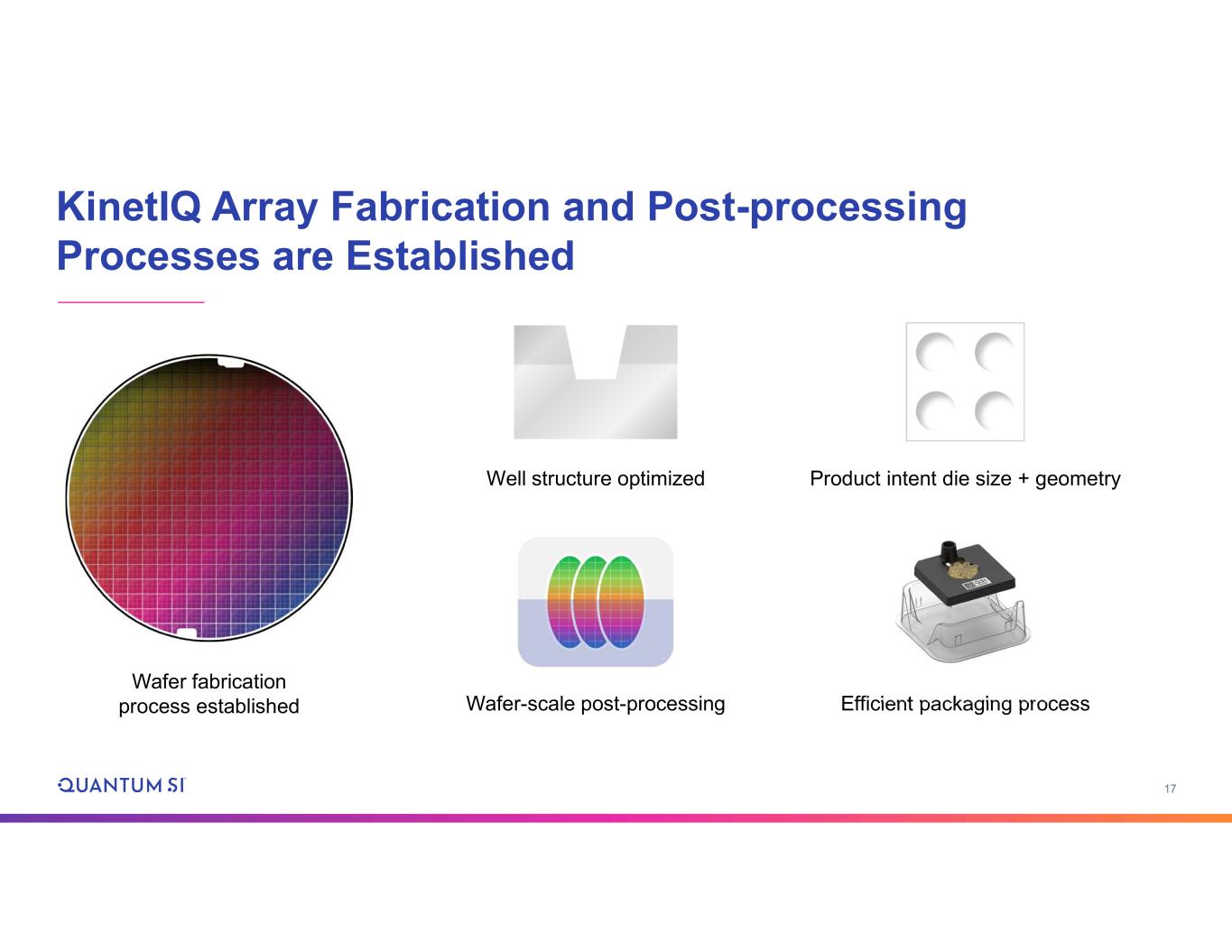
17 KinetIQ Array Fabrication and Post-processing Processes are Established Wafer fabrication process established Well structure optimized Efficient packaging processWafer-scale post-processing Product intent die size + geometry
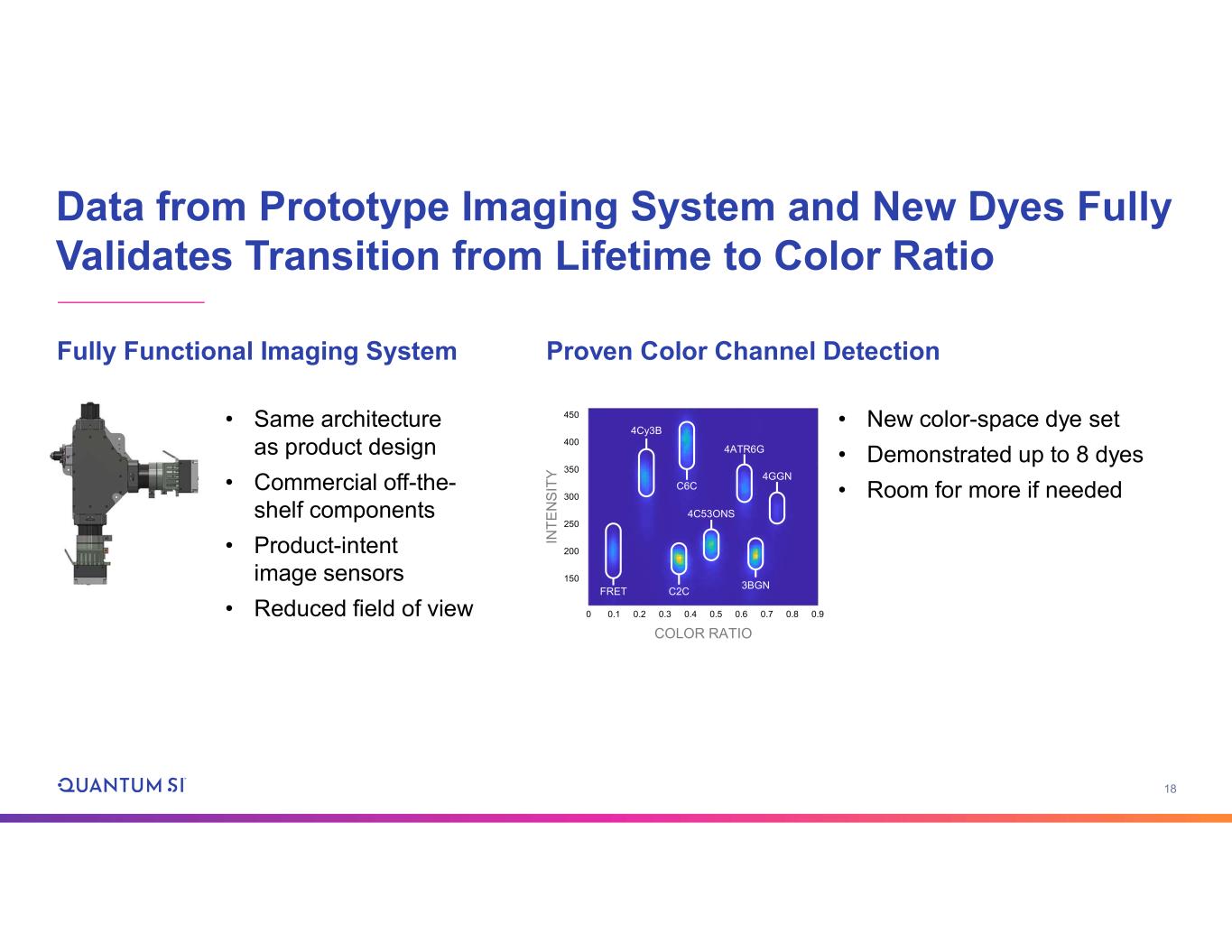
18 Data from Prototype Imaging System and New Dyes Fully Validates Transition from Lifetime to Color Ratio • Same architecture as product design • Commercial off-the- shelf components • Product-intent image sensors • Reduced field of view • New color-space dye set • Demonstrated up to 8 dyes • Room for more if needed 450 0 400 350 300 250 200 150 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 IN T E N S IT Y COLOR RATIO 4Cy3B C6C 4ATR6G 4GGN C2CFRET 3BGN 4C53ONS Fully Functional Imaging System Proven Color Channel Detection
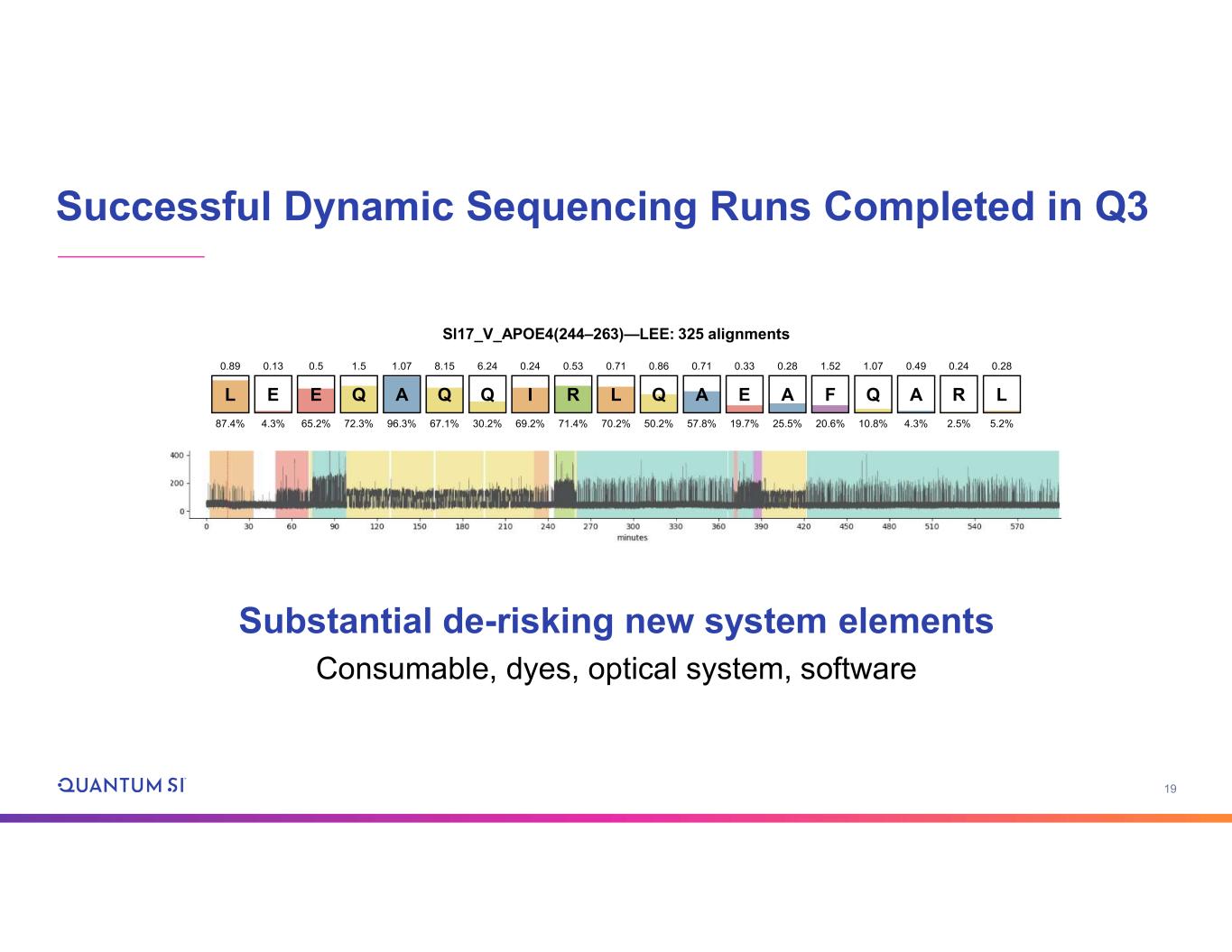
19 Successful Dynamic Sequencing Runs Completed in Q3 Substantial de-risking new system elements Consumable, dyes, optical system, software L 87.4% 0.89 E 65.2% 0.5 Q 72.3% 1.5 A 96.3% 1.07 Q 67.1% 8.15 Q 30.2% 6.24 I 69.2% 0.24 R 71.4% 0.53 L 70.2% 0.71 Q 50.2% 0.86 A 57.8% 0.71 E 19.7% 0.33 A 25.5% 0.28 F 20.6% 1.52 Q 10.8% 1.07 A 4.3% 0.49 R 2.5% 0.24 L 5.2% 0.28 4.3% E 0.13 Sl17_V_APOE4(244–263)—LEE: 325 alignments
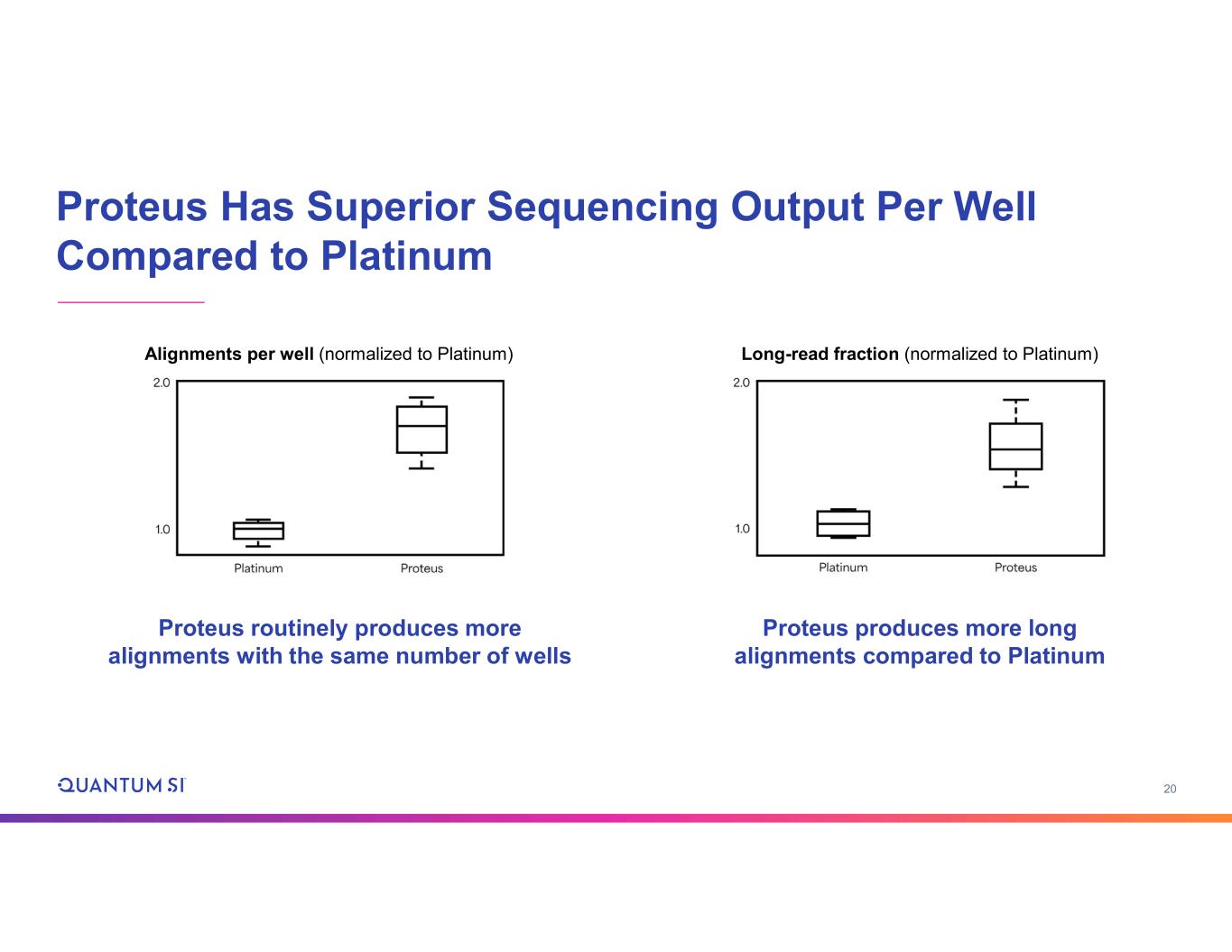
20 Proteus Has Superior Sequencing Output Per Well Compared to Platinum Proteus routinely produces more alignments with the same number of wells Proteus produces more long alignments compared to Platinum Alignments per well (normalized to Platinum) Long-read fraction (normalized to Platinum)
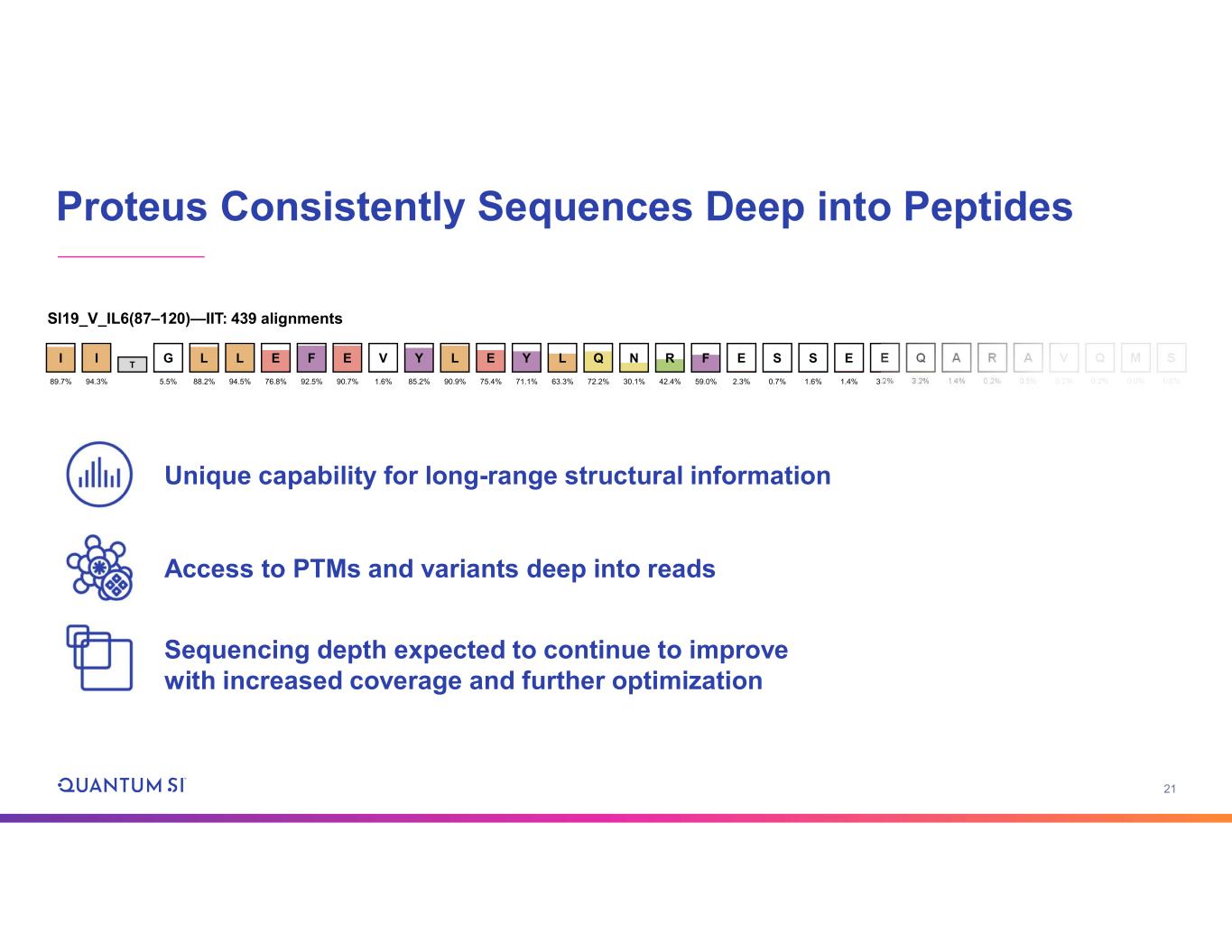
21 Proteus Consistently Sequences Deep into Peptides Unique capability for long-range structural information Access to PTMs and variants deep into reads Sequencing depth expected to continue to improve with increased coverage and further optimization Sl19_V_IL6(87–120)—IIT: 439 alignments 30.1% I 89.7% E 2.3% G 5.5% L 88.2% L 94.5% E 76.8% F 92.5% E 90.7% V 1.6% Y 85.2% L 90.9% E 75.4% Y 71.1% L 63.3% Q 72.2% NI 94.3% T R 42.4% F 59.0% S 0.7% S 1.6% E 1.4% E 3.2% Q 3.2% A 1.4% R 0.2% A 0.5% V 0.2% Q 0.2% M 0.0% S 0.0%
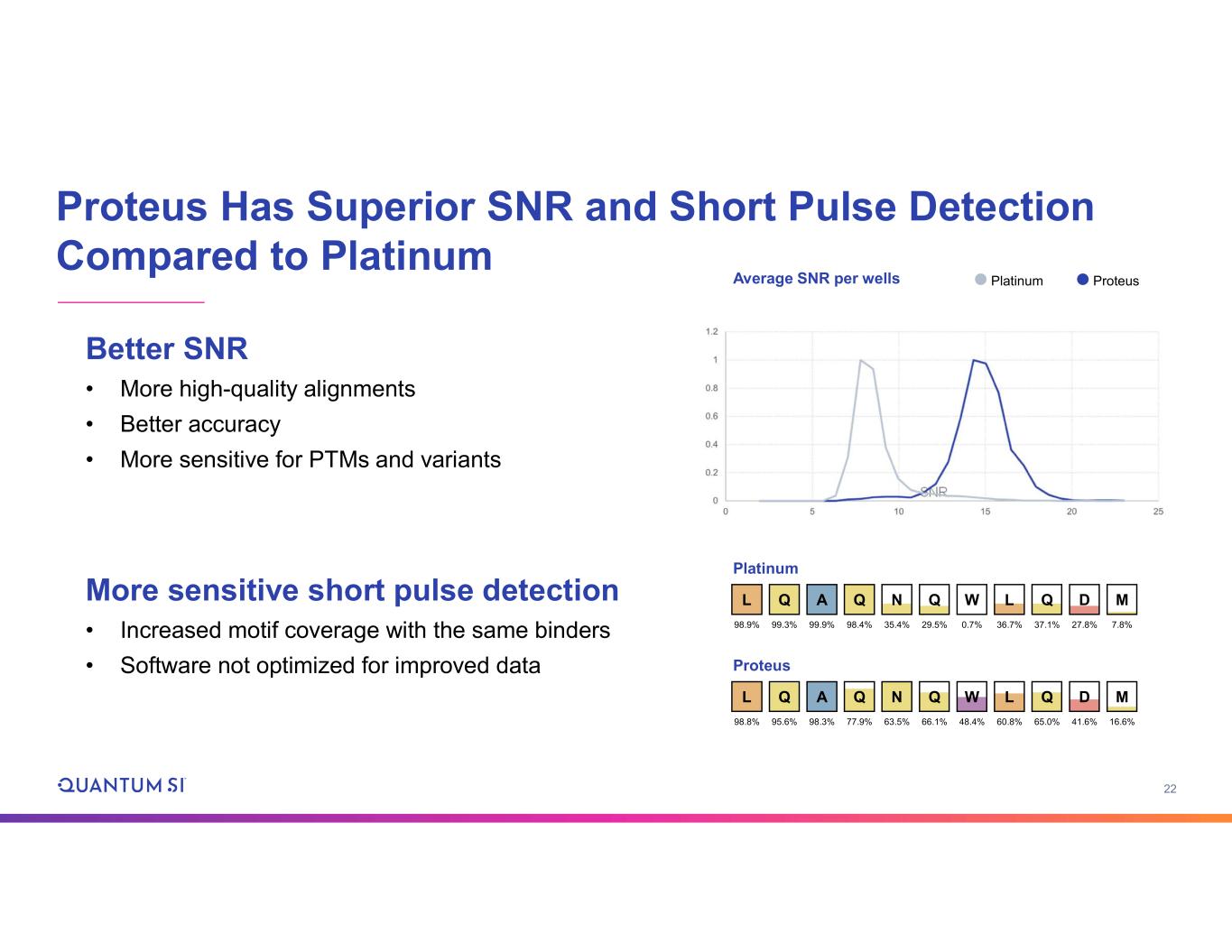
22 Proteus Has Superior SNR and Short Pulse Detection Compared to Platinum Better SNR • More high-quality alignments • Better accuracy • More sensitive for PTMs and variants More sensitive short pulse detection • Increased motif coverage with the same binders • Software not optimized for improved data Proteus L 98.8% Q 95.6% A 98.3% M 16.6% Q 77.9% N 63.5% Q 66.1% W 48.4% L 60.8% Q 65.0% D 41.6% Platinum L 98.9% Q 99.3% A 99.9% M 7.8% Q 98.4% N 35.4% Q 29.5% W 0.7% L 36.7% Q 37.1% D 27.8% Platinum ProteusAverage SNR per wells SNR
23 Proteus Sequencing Performance is on Track to Far Exceed Platinum at Launch Proteus data is already superior to Platinum data Data generated to date is without optimization Current algorithms | Current V4 binder set | Current cutters and chemistry Additional improvements expected before launch

24 Integrated System in Development First Integrated System Expected 1Q26 • Full integration of imaging and workflow automation • Product-like consumables • Full field of view supporting 20M wells
25 Proteus Program Summary Transition to new architecture is proven — team is executing towards product launch for YE2026 Proteus architecture shows significant improvement over Platinum in the quantity and quality of sequencing output Platform launch is aligned with improvements in biochemistry and library prep that will drive additional performance gains

26 Long-term Technology Roadmap
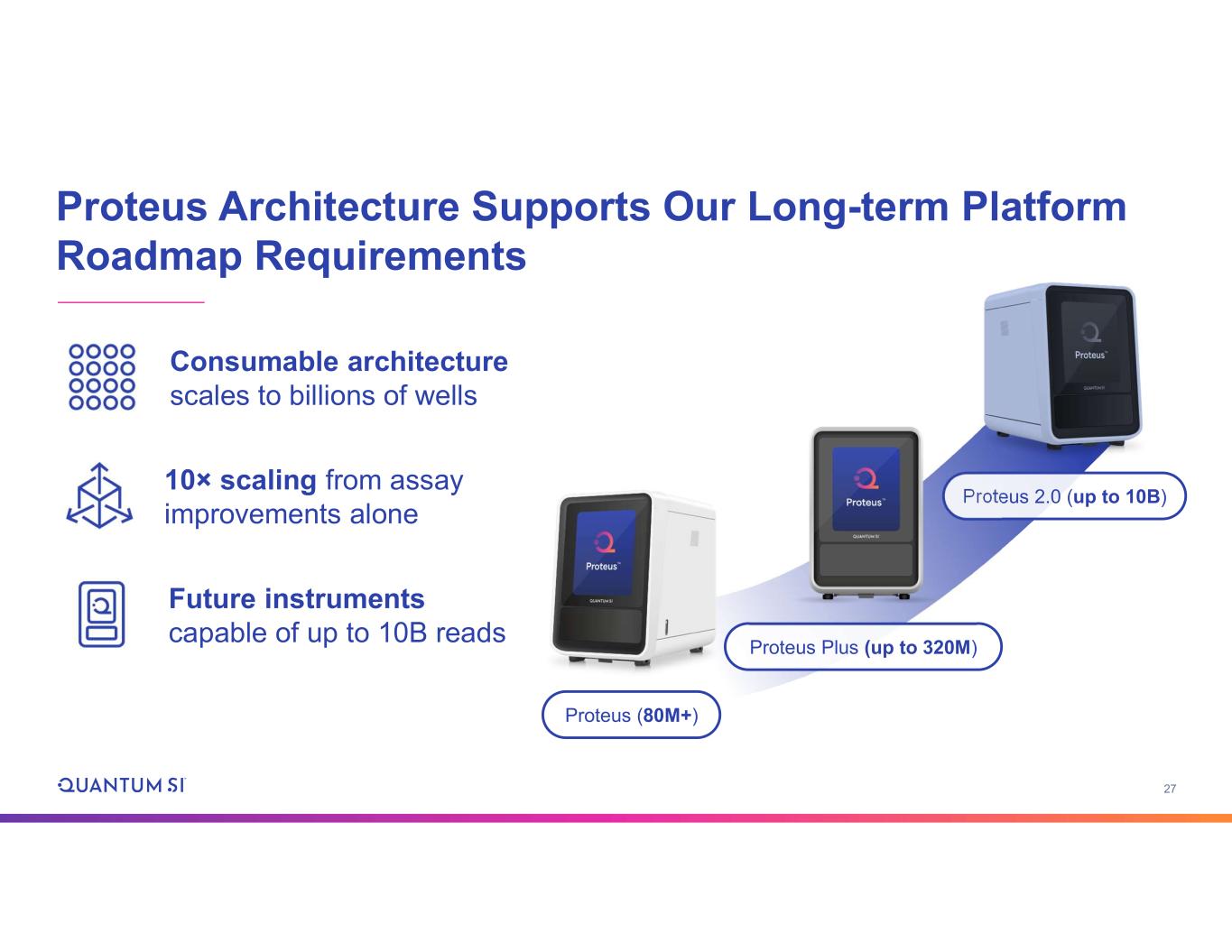
27 Proteus Architecture Supports Our Long-term Platform Roadmap Requirements Proteus 2.0 (up to 10B) Proteus (80M+) Proteus Plus (up to 320M) Consumable architecture scales to billions of wells 10× scaling from assay improvements alone Future instruments capable of up to 10B reads
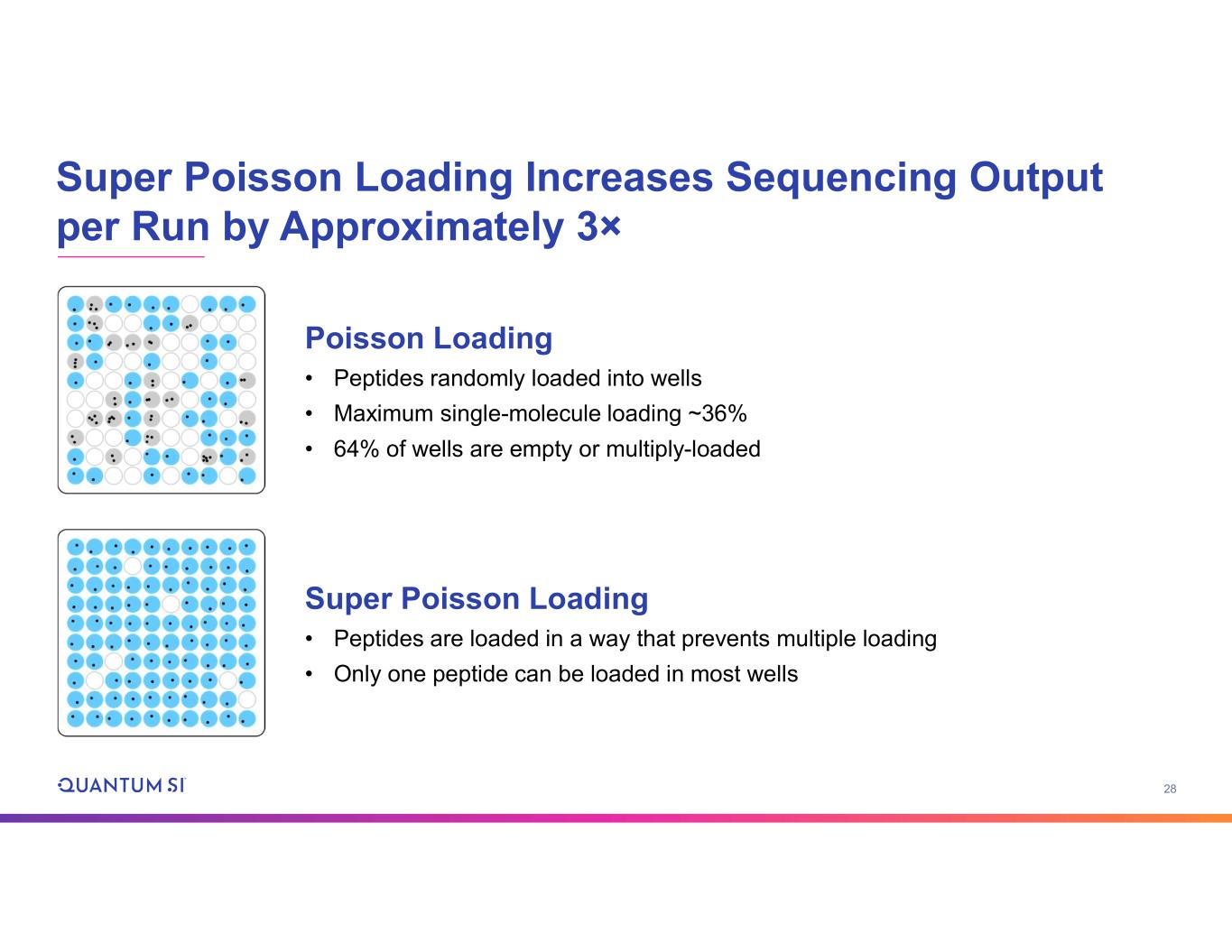
28 Super Poisson Loading Increases Sequencing Output per Run by Approximately 3× Super Poisson Loading • Peptides are loaded in a way that prevents multiple loading • Only one peptide can be loaded in most wells Poisson Loading • Peptides randomly loaded into wells • Maximum single-molecule loading ~36% • 64% of wells are empty or multiply-loaded
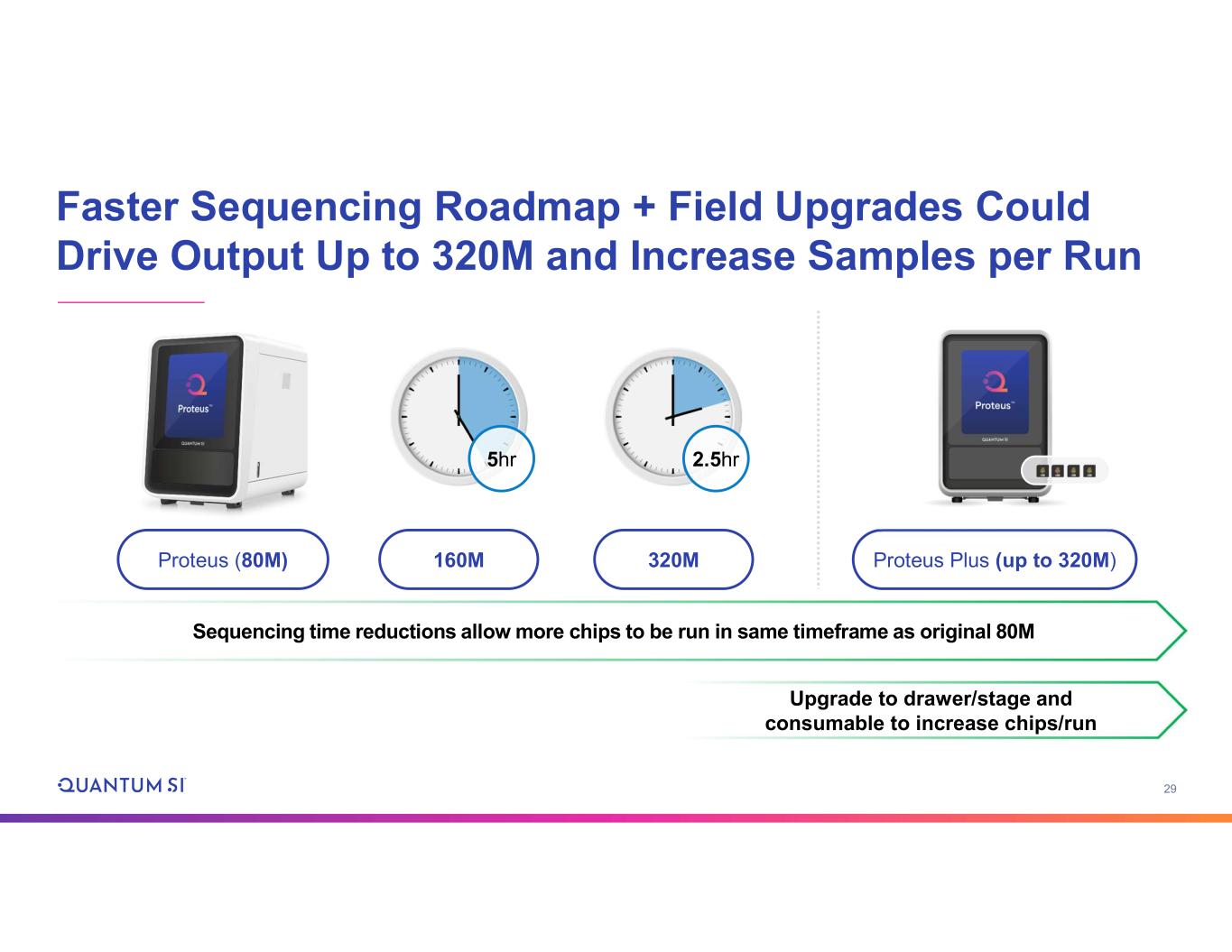
29 Faster Sequencing Roadmap + Field Upgrades Could Drive Output Up to 320M and Increase Samples per Run Proteus (80M) Sequencing time reductions allow more chips to be run in same timeframe as original 80M Upgrade to drawer/stage and consumable to increase chips/run Proteus Plus (up to 320M)160M 320M 5hr 2.5hr
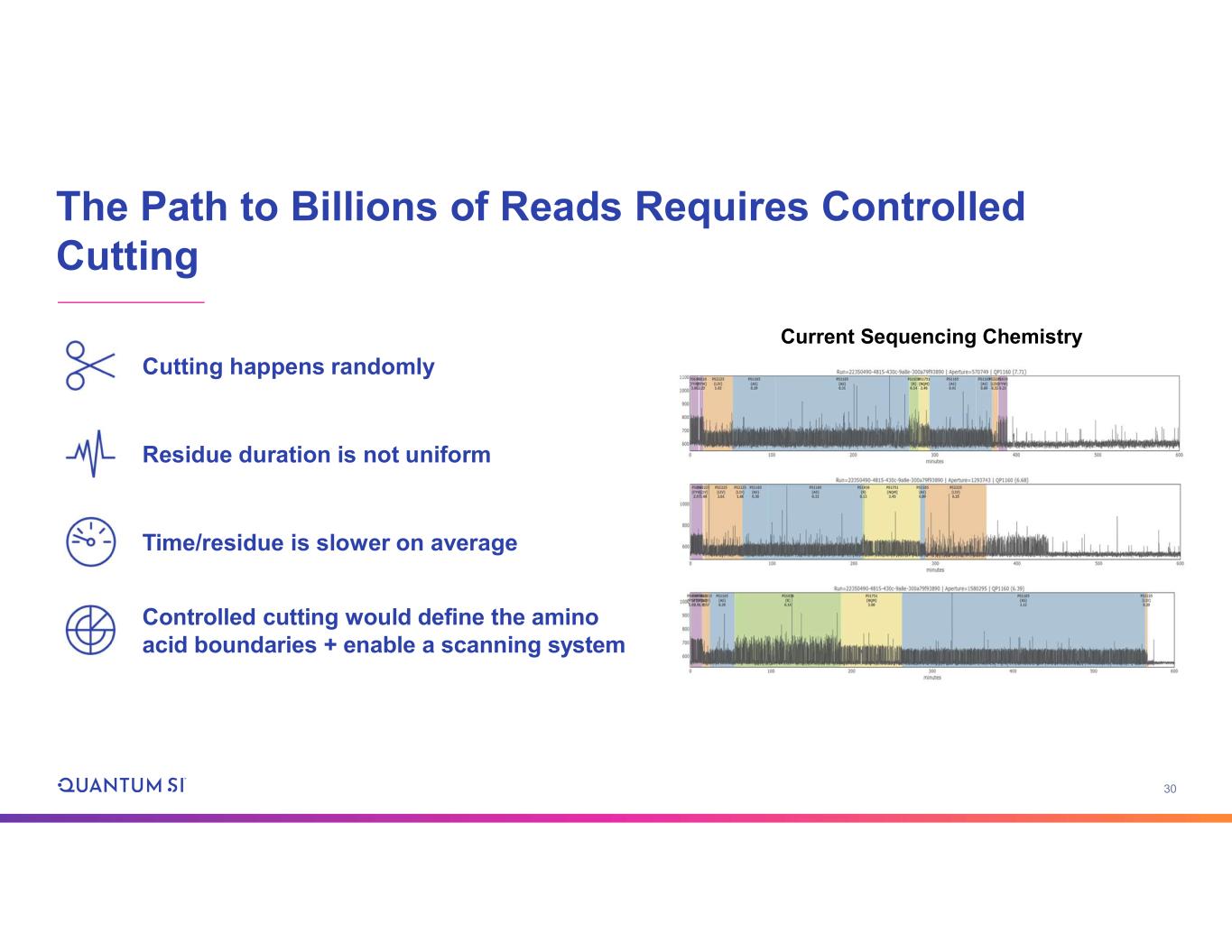
30 The Path to Billions of Reads Requires Controlled Cutting Current Sequencing Chemistry Cutting happens randomly Residue duration is not uniform Time/residue is slower on average Controlled cutting would define the amino acid boundaries + enable a scanning system
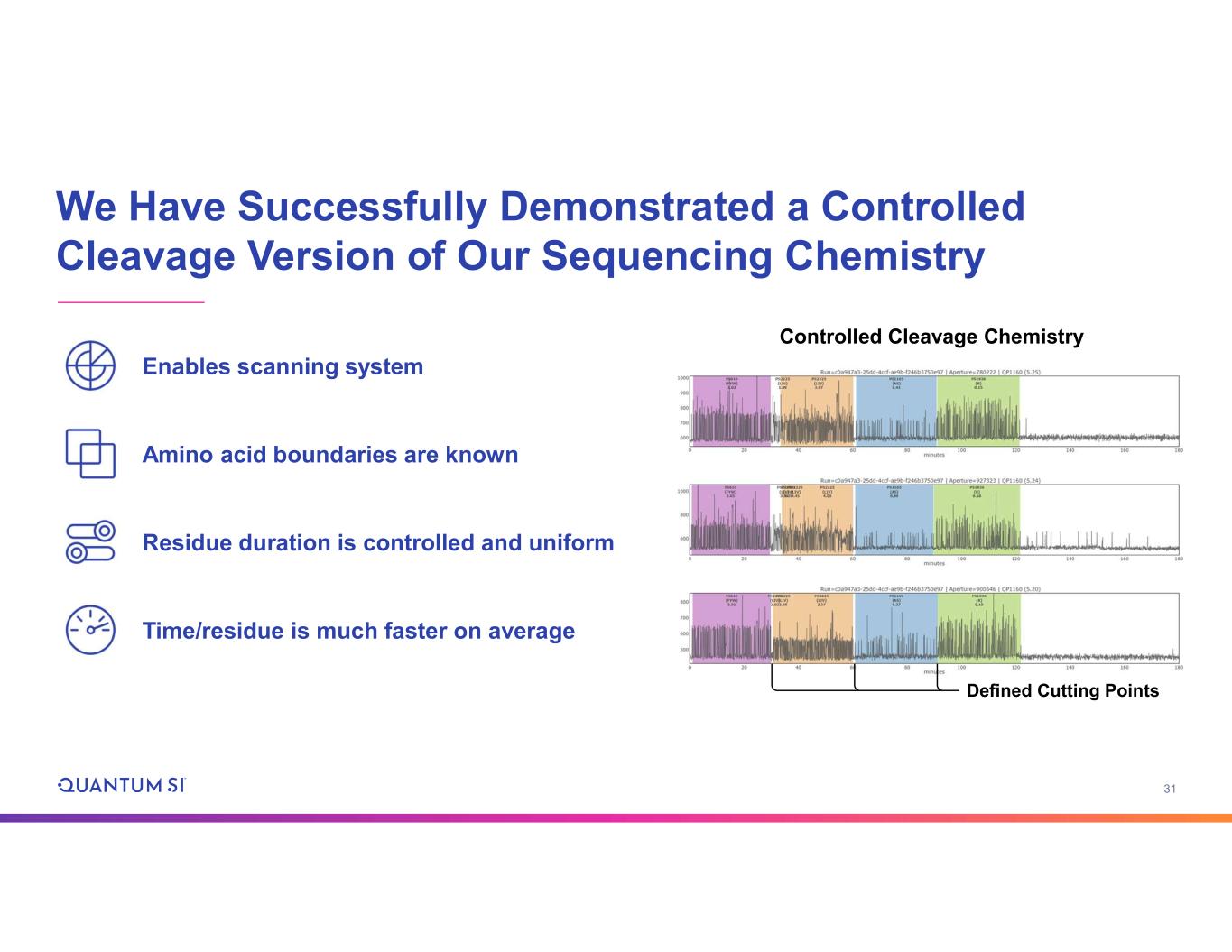
31 We Have Successfully Demonstrated a Controlled Cleavage Version of Our Sequencing Chemistry Enables scanning system Amino acid boundaries are known Residue duration is controlled and uniform Time/residue is much faster on average Controlled Cleavage Chemistry Defined Cutting Points

32 Proteus 2.0 can incorporate high-speed scanning and microfluidic delivery system Scanning system enables billions of reads Multiple imaging locations within one flow cell Efficient usage of device area Enabled by controlled cleavage chemistry Combining Controlled Cleavage with High-speed Scanning Scales the Architecture to Billions of Reads Multiple imaging regions per flow cell

33 The Path to Detecting all 20 Amino Acids John Vieceli, Chief Product Officer November 19, 2025

34 Develop a set of N-terminal amino acid recognizers for twenty canonical amino acids on Proteus Vision

Recognizer Roadmap
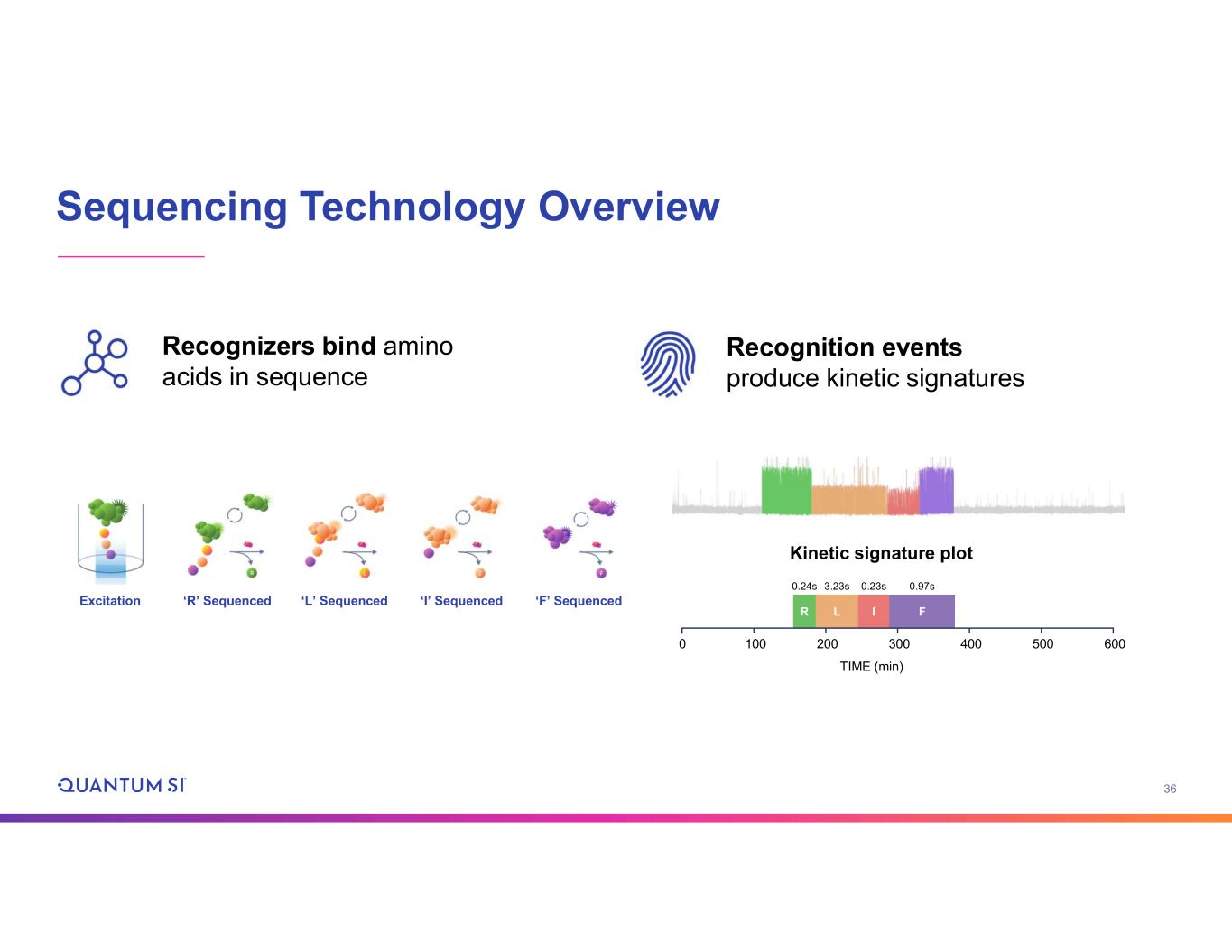
36 Sequencing Technology Overview Recognizers bind amino acids in sequence Recognition events produce kinetic signatures R L I F 6005004003002001000 0.24s 3.23s 0.23s 0.97s TIME (min) Kinetic signature plot Excitation ‘R’ Sequenced ‘L’ Sequenced ‘I’ Sequenced ‘F’ Sequenced
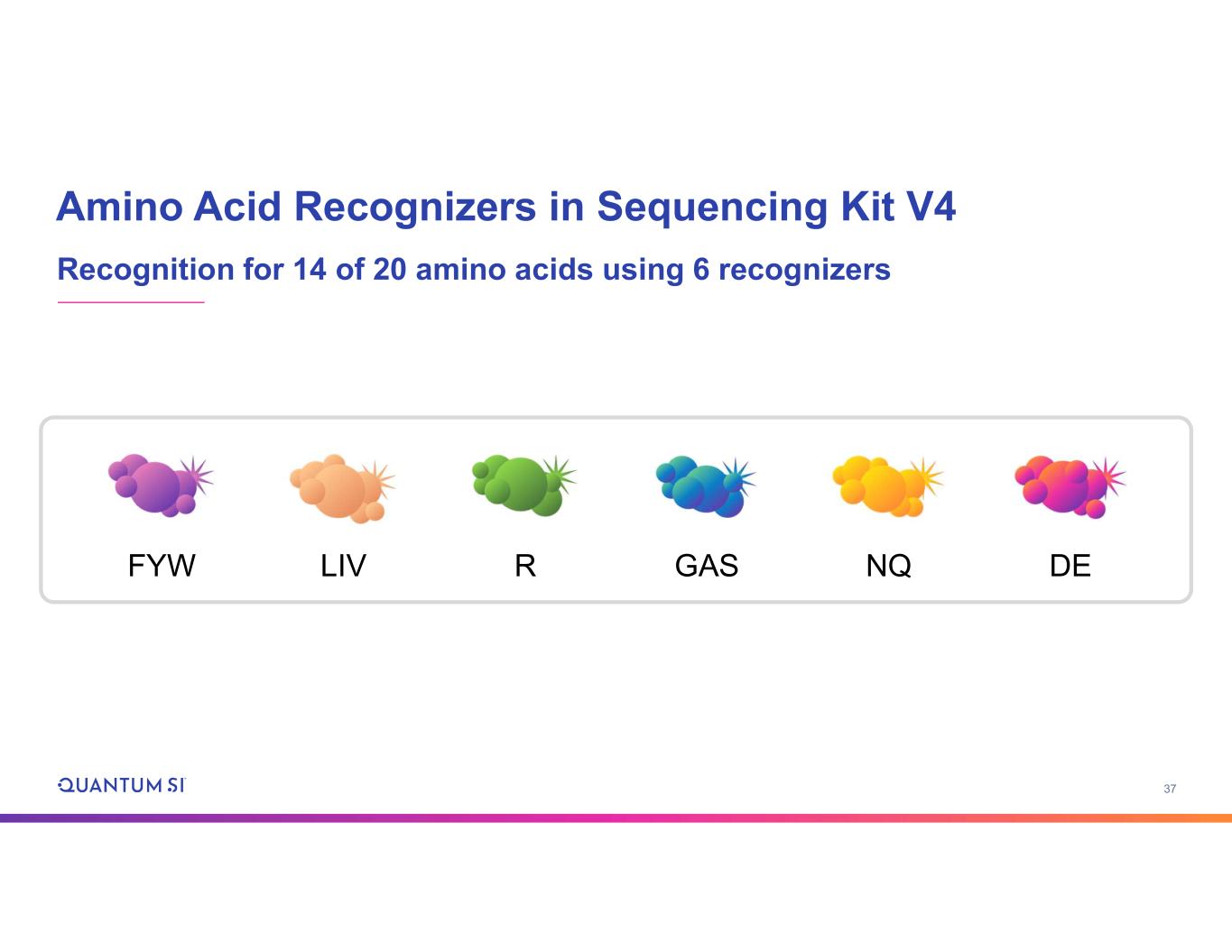
37 Amino Acid Recognizers in Sequencing Kit V4 Recognition for 14 of 20 amino acids using 6 recognizers DEGASFYW LIV R NQ
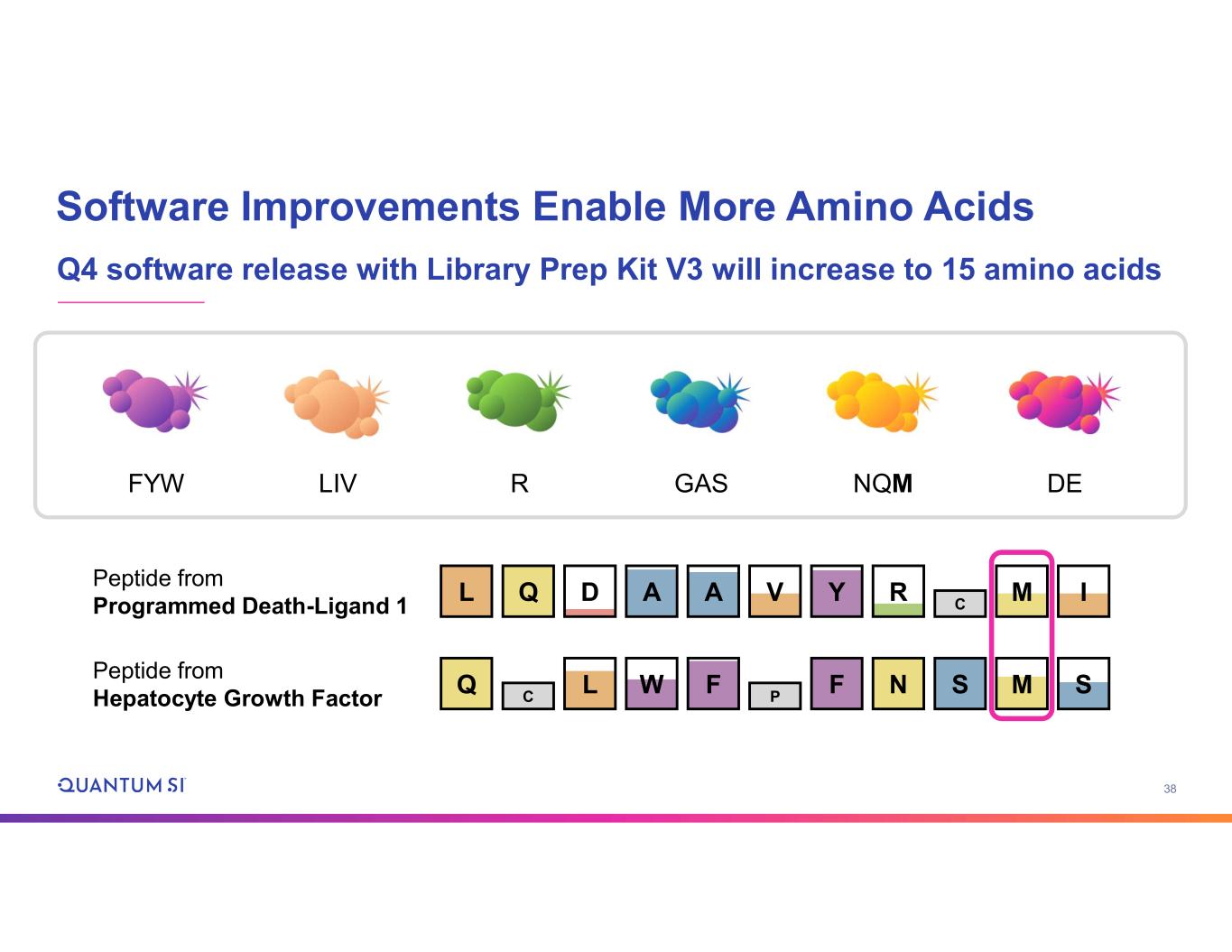
38 Software Improvements Enable More Amino Acids Q4 software release with Library Prep Kit V3 will increase to 15 amino acids DEGASFYW LIV R NQM Peptide from Programmed Death-Ligand 1 Peptide from Hepatocyte Growth Factor DL Q A A V Y R C M I Q F NC M SL W F P S
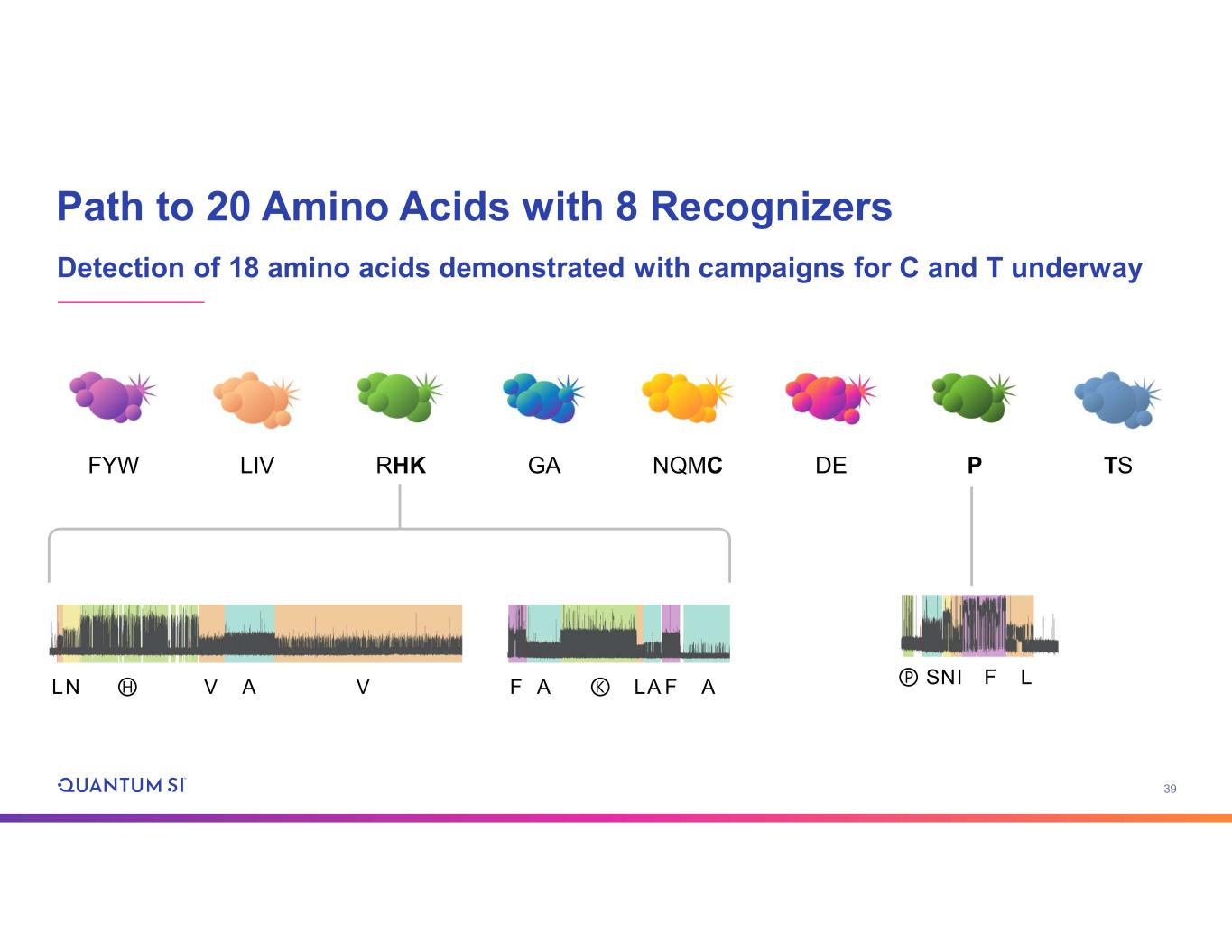
39 Path to 20 Amino Acids with 8 Recognizers Detection of 18 amino acids demonstrated with campaigns for C and T underway DEGAFYW LIV P TSRHK NQMC LN Ⓗ V A V F A Ⓚ LA AF SⓅ NI LF
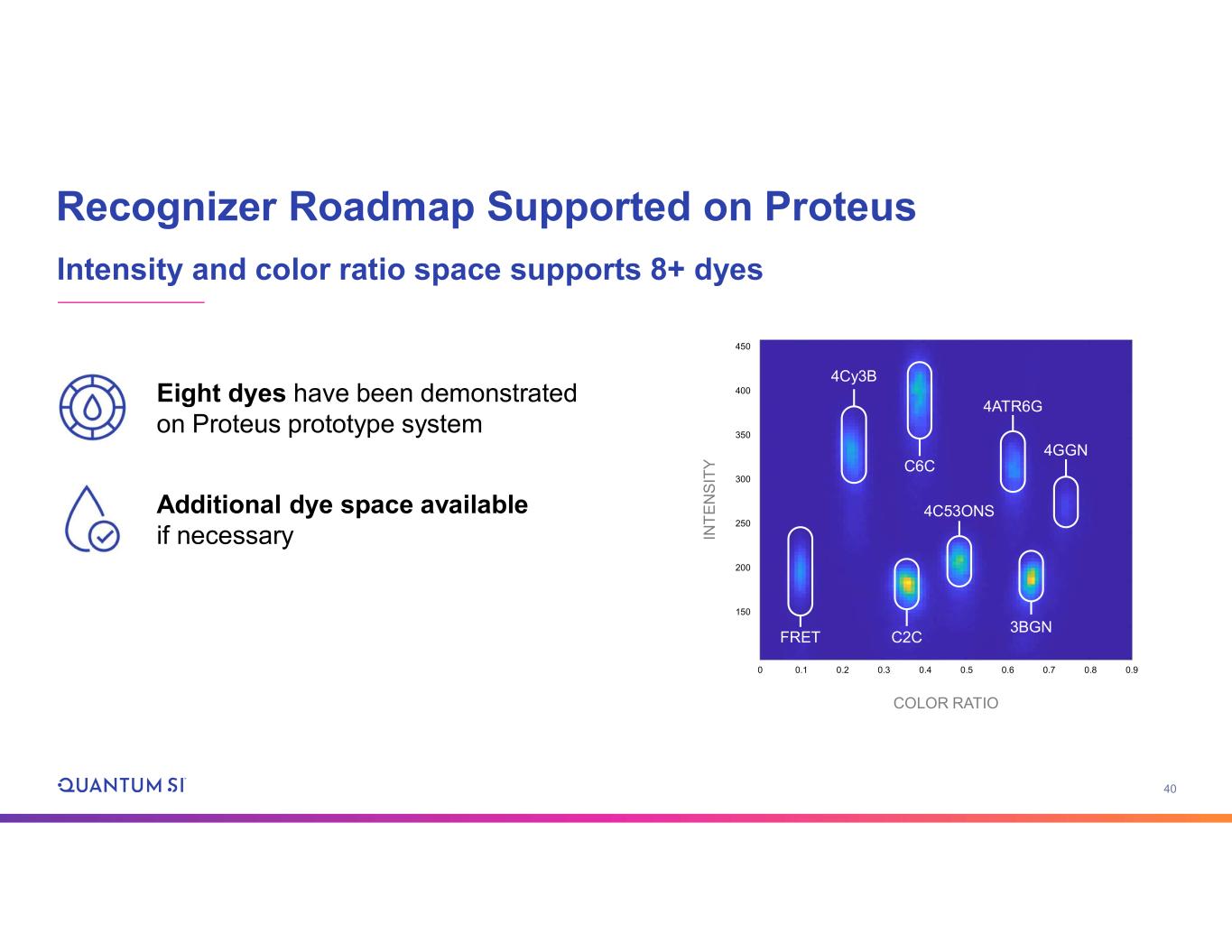
40 450 0 400 350 300 250 200 150 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 IN T E N S IT Y COLOR RATIO Recognizer Roadmap Supported on Proteus Intensity and color ratio space supports 8+ dyes 4Cy3B C6C 4ATR6G 4GGN C2CFRET 3BGN 4C53ONS Eight dyes have been demonstrated on Proteus prototype system Additional dye space available if necessary

Recognizer Development Pipeline
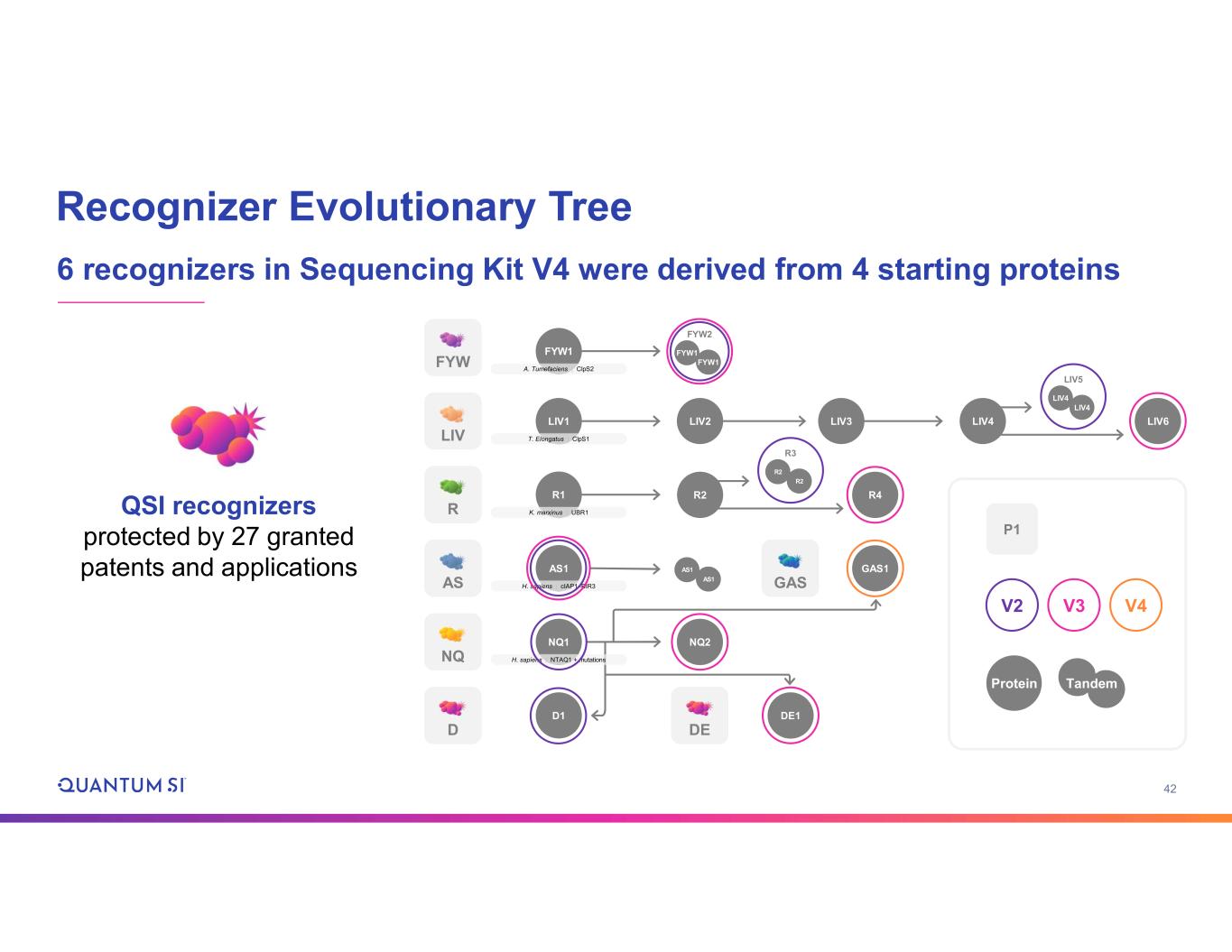
42 Recognizer Evolutionary Tree 6 recognizers in Sequencing Kit V4 were derived from 4 starting proteins FYW LIV GASAS R NQ D DE FYW2 FYW1 FYW1FYW1 A. Tumefaciens | ClpS2 LIV1 T. Elongatus | ClpS1 R1 K. marxinus | UBR1 AS1 H. sapiens | cIAP1–BIR3 NQ1 H. sapiens | NTAQ1 + mutations D1 LIV2 LIV3 LIV4 LIV5 LIV4 LIV4 LIV6 R2 R3 R2 R2 R4 AS1 AS1 GAS1 NQ2 DE1 P1 V2 V3 V4 Protein Tandem QSI recognizers protected by 27 granted patents and applications
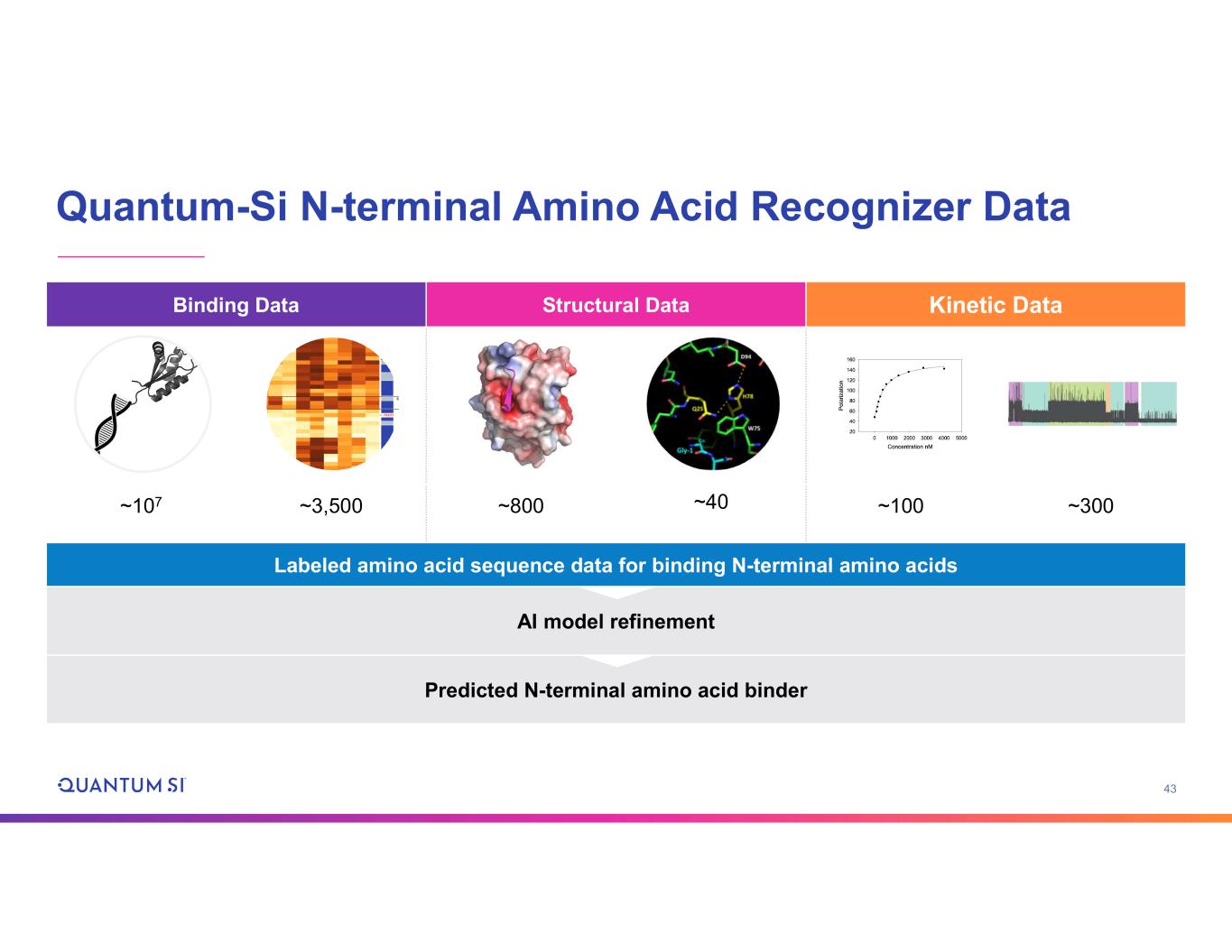
43 Quantum-Si N-terminal Amino Acid Recognizer Data Kinetic DataStructural DataBinding Data ~300~100~40~800~3,500~107 Labeled amino acid sequence data for binding N-terminal amino acids AI model refinement Predicted N-terminal amino acid binder
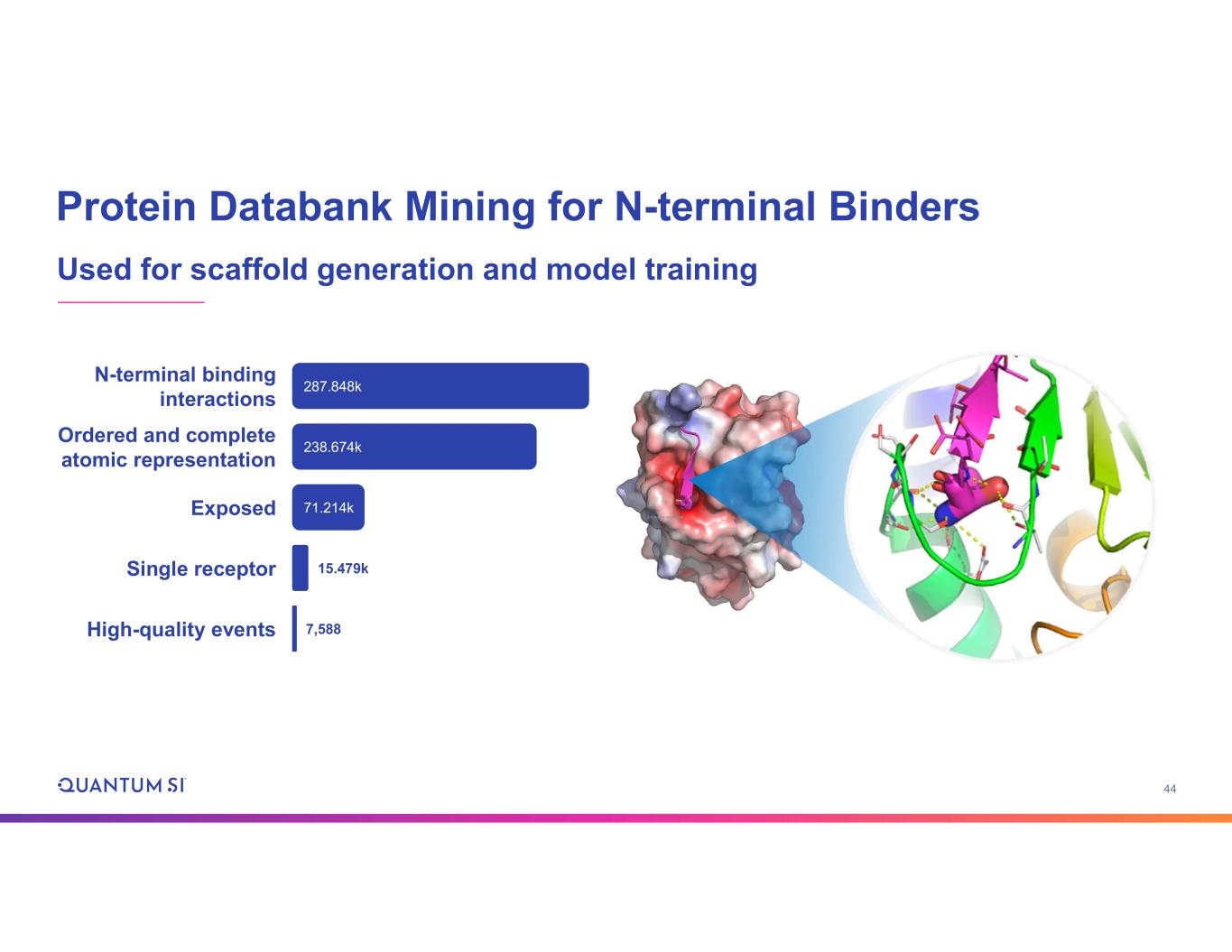
44 Protein Databank Mining for N-terminal Binders Used for scaffold generation and model training High-quality events 7,588 71.214kExposed Single receptor 15.479k 238.674k Ordered and complete atomic representation 287.848k N-terminal binding interactions
45 AI Model Refinement Enabled By Academia and Industry Ligand message- passing neural network Evolutionary scale modeling GPU hardware acceleration
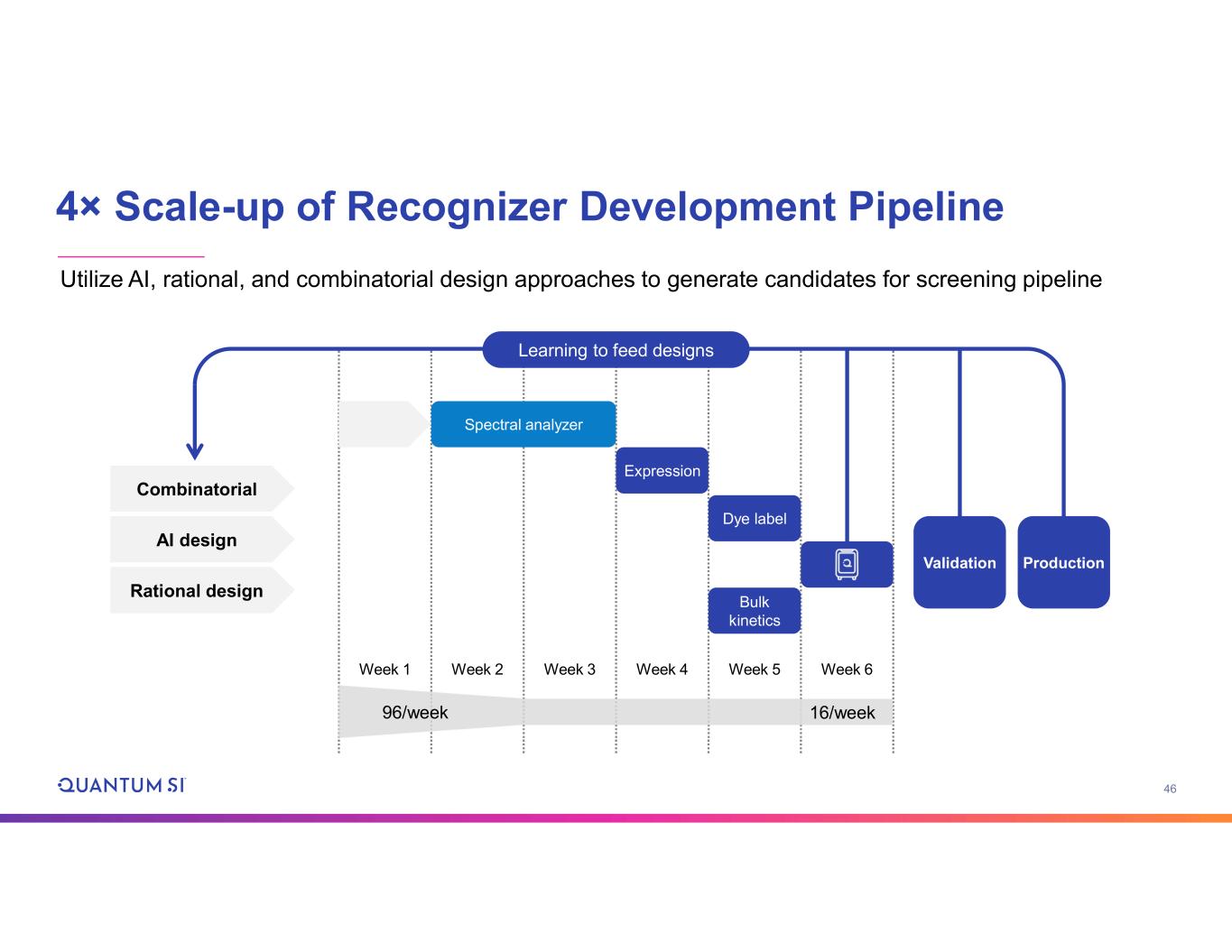
46 4× Scale-up of Recognizer Development Pipeline Utilize AI, rational, and combinatorial design approaches to generate candidates for screening pipeline Week 1 Week 2 Week 3 Week 4 Week 5 Week 6 Spectral analyzer Expression Dye label Bulk kinetics 96/week 16/week Validation Production Combinatorial AI design Rational design Learning to feed designs
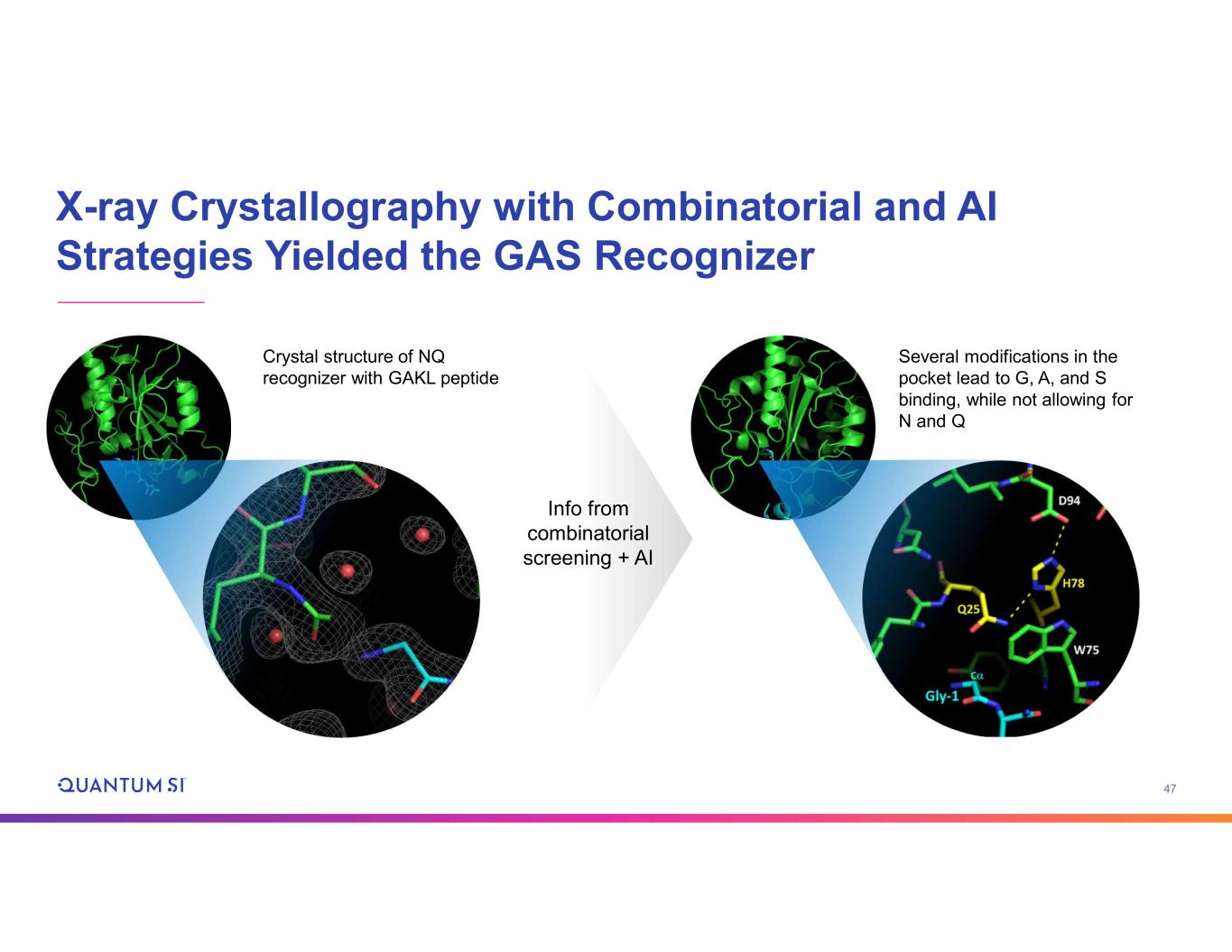
47 X-ray Crystallography with Combinatorial and AI Strategies Yielded the GAS Recognizer Crystal structure of NQ recognizer with GAKL peptide Several modifications in the pocket lead to G, A, and S binding, while not allowing for N and Q Info from combinatorial screening + AI
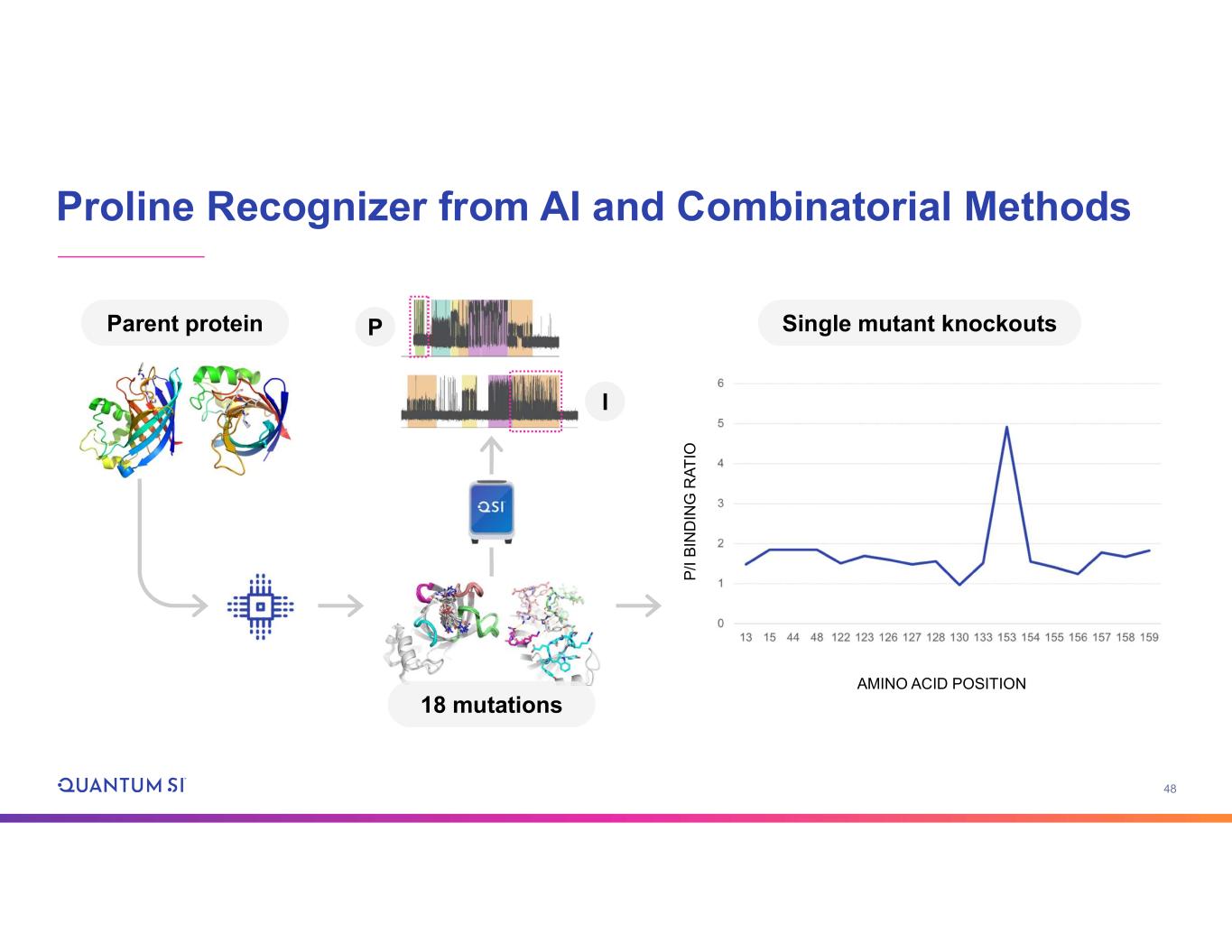
48 Proline Recognizer from AI and Combinatorial Methods Parent protein 18 mutations P I Single mutant knockouts P /I B IN D IN G R A T IO AMINO ACID POSITION
49 Summary of the Path to Twenty Amino Acids Recognizer development program is being scaled 4× to achieve detection of all canonical amino acids in 2026 and release on Proteus in 2027 Efforts in artificial intelligence utilize Quantum-Si screening data to refine deep learning and language models, in conjunction with rational and combinatorial design, to accelerate the pace of recognizer development Current recognizer pipeline has candidates to go from 14 amino acids with Sequencing Kit V4 to 18 at Proteus launch in 2026

50 PTM Analysis Solutions Brian Reed, PhD Head of Research
51 Agenda PTM Solutions on Proteus Advantages of single-molecule detection for PTMs1 PTM applications on the path to Proteus launch3 Broad access to PTMs on Proteus2
52 Su P G Me Methylation (Lys/Arg) Phosphorylation (Ser/Thr/Tyr) Palmitoylation (Cys) Sumoylation (Lys) Ubiquitination (Lys) Citrullination (Arg) AC Pal Ub Glycosylation (Ser/Thr/Asn) Acetylation (Lys) Translation Cit A post-translational modification (PTM) is a chemical change to an amino acid made after a protein has been expressed. There are hundreds of different types of PTMs. A handful are of high interest in the proteomics due to their high abundance and roles in disease pathways. Examples: phosphorylation, glycosylation, ubiquitination. PTMs determine actual protein function — making them essential for understanding disease mechanisms and therapeutic targets — but have been difficult to study with current proteomics technology. PTMs and Proteoforms are the Next Frontier in Human Health

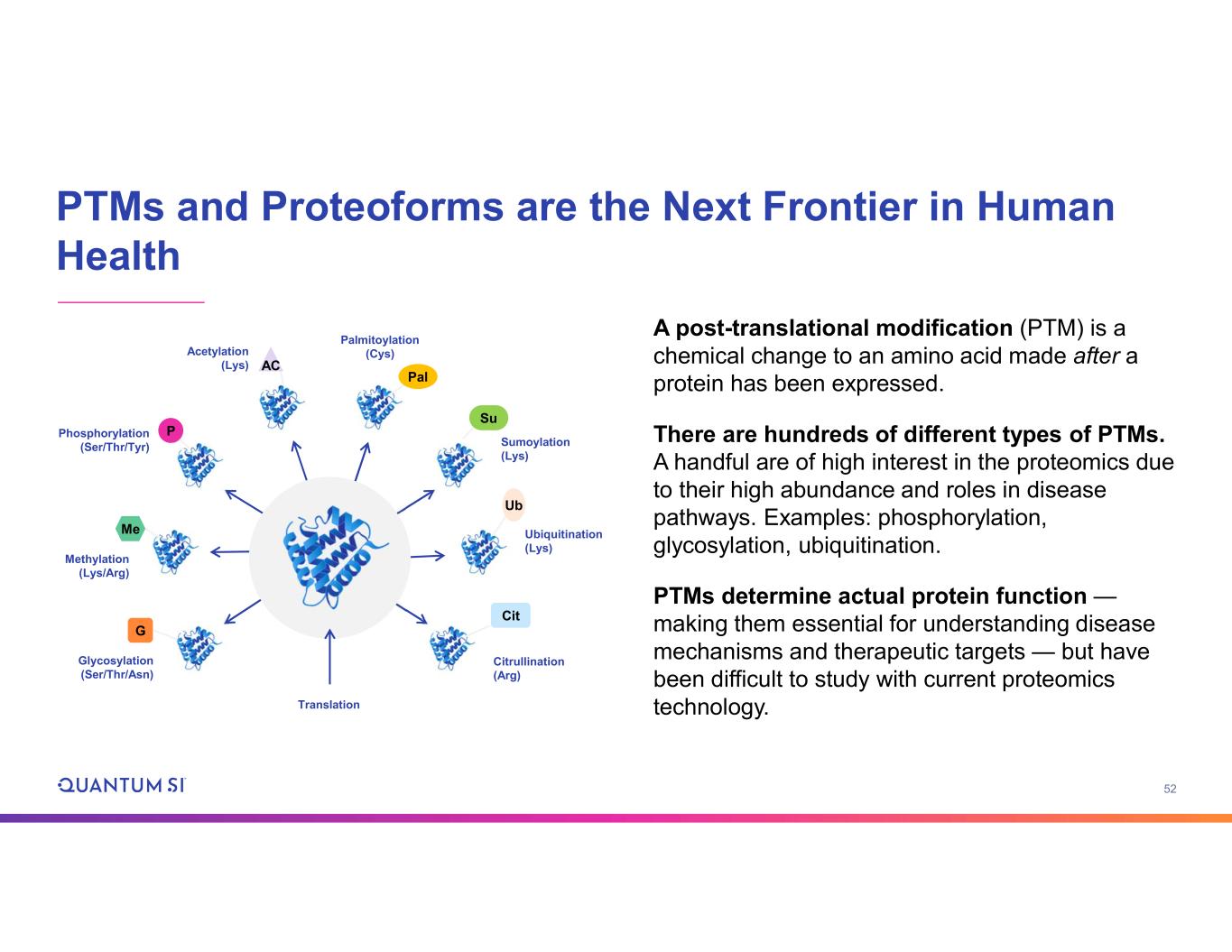
53 Challenge to Detecting PTMs With Existing Technologies Mass spectrometry • Quantification difficult • Ambiguity in modification site assignment • Multi-site combinations extremely challenging • Workflow and analytical complexity (limits clinical adoption) Affinity-based platforms • Detect proteins but provide no proteoform information OR • Need costly site-specific affinity reagents for every protein
54 Advantages of Single-molecule Detection for PTMs • Quantitation straightforward due to direct counting of molecules • Direct PTM site identification • Access to multi-site PTM combinations • Simple workflows for broad PTM proteome coverage; clinical compatibility • Universal PTM detection reagents and assays for all proteins
55 Single-molecule Detection Enables Broad PTM Coverage NGPS unlocks broad and universal access to PTMs with three complimentary approaches | Can be used in combination to address the most challenging needs in PTM detection Kinetics PTM detected via kinetic changes at upstream AAs Pre-recognition PTM detected with a PTM binder prior to sequencing Direct NAA detection PTM detected with an N-terminal PTM binder during sequencing Complete AA coverage and deep peptide sequencing will enable very high proteome-wide PTM coverage on Proteus for all three methods
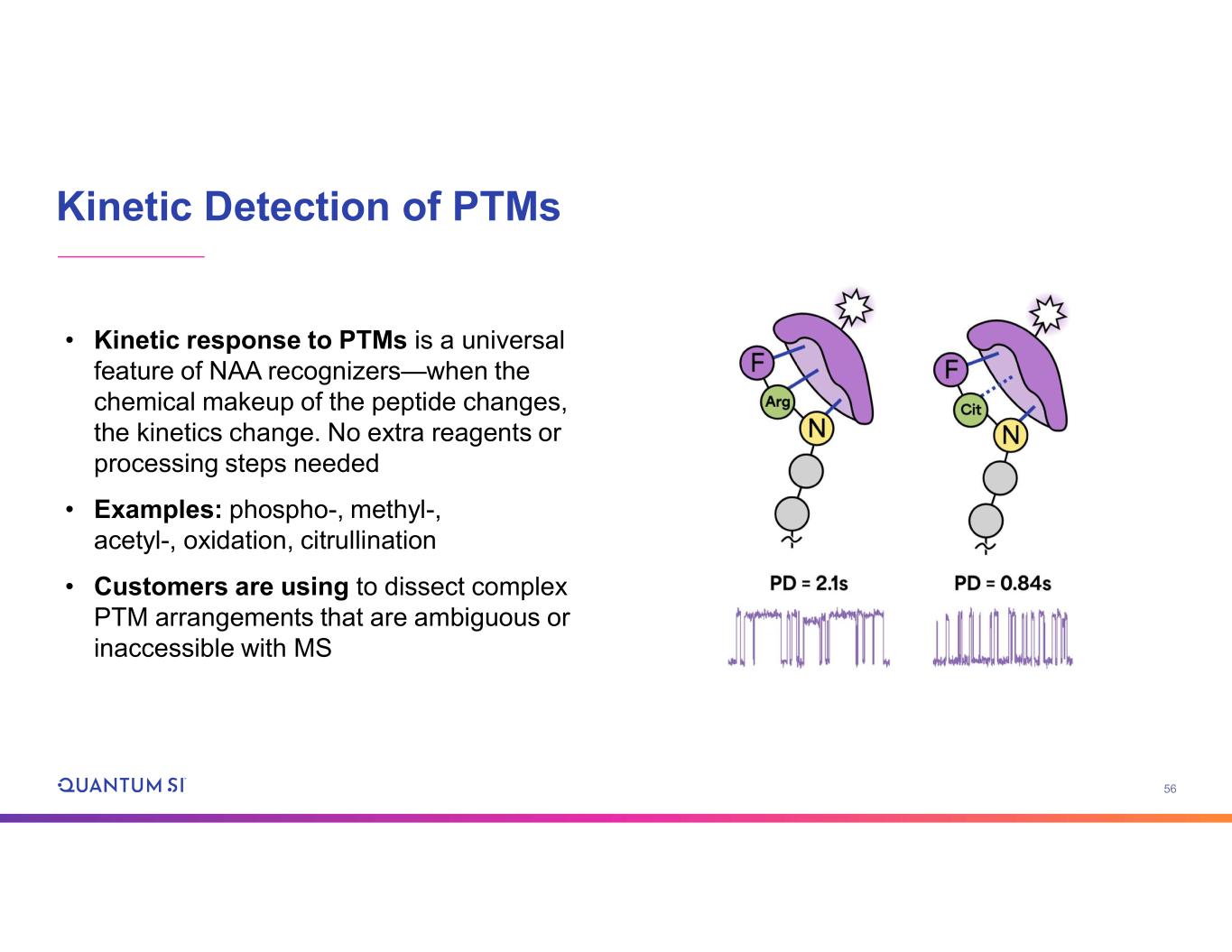
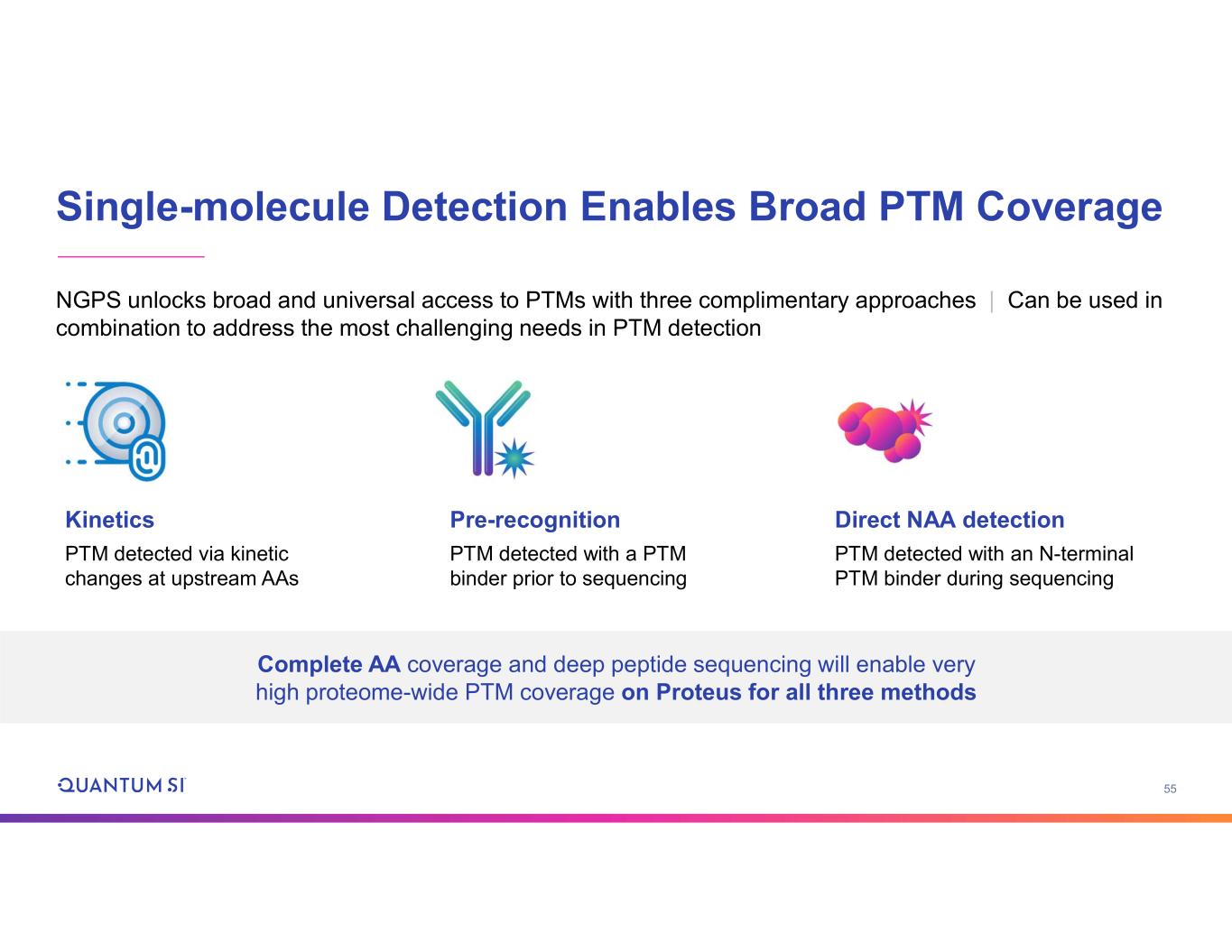
56 Kinetic Detection of PTMs • Kinetic response to PTMs is a universal feature of NAA recognizers—when the chemical makeup of the peptide changes, the kinetics change. No extra reagents or processing steps needed • Examples: phospho-, methyl-, acetyl-, oxidation, citrullination • Customers are using to dissect complex PTM arrangements that are ambiguous or inaccessible with MS
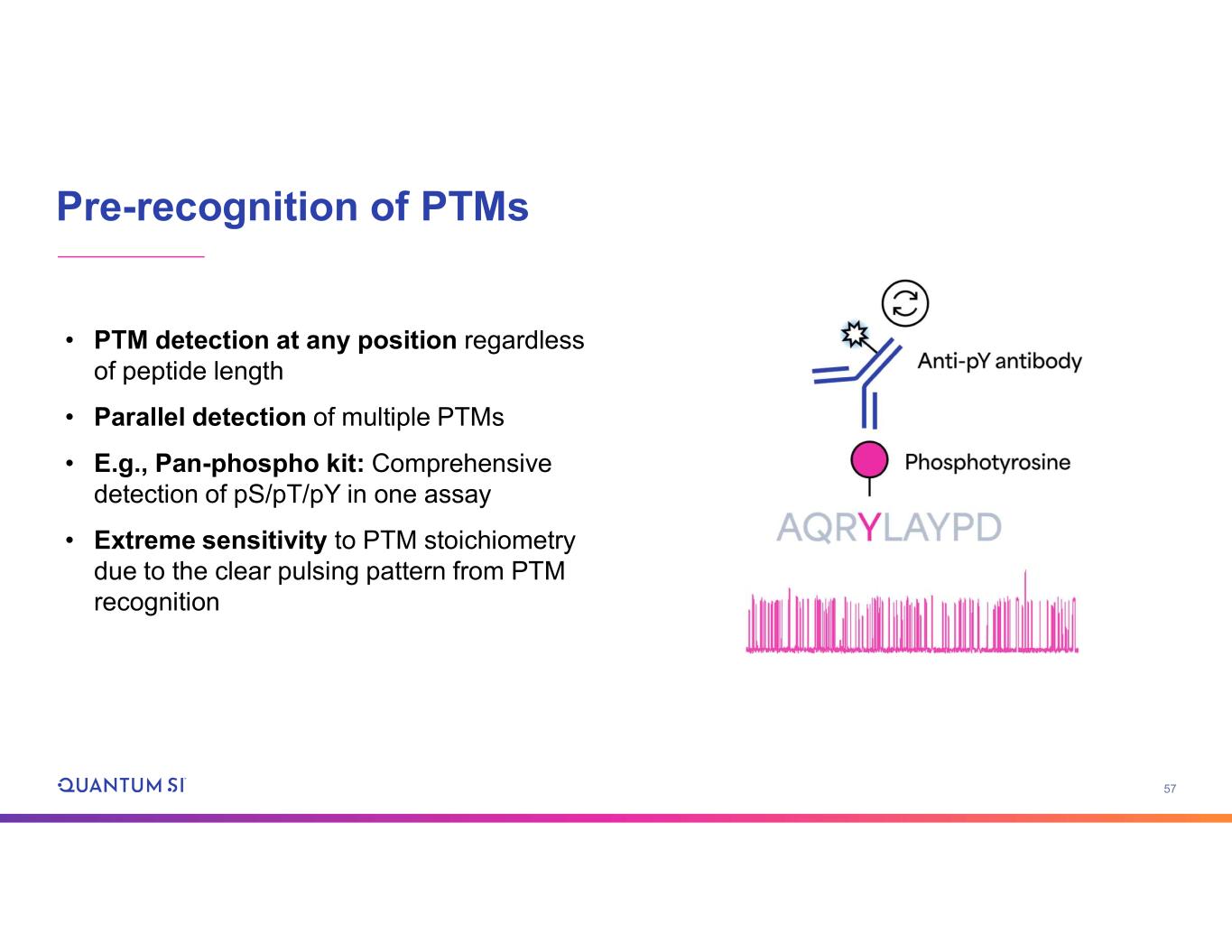
57 Pre-recognition of PTMs • PTM detection at any position regardless of peptide length • Parallel detection of multiple PTMs • E.g., Pan-phospho kit: Comprehensive detection of pS/pT/pY in one assay • Extreme sensitivity to PTM stoichiometry due to the clear pulsing pattern from PTM recognition
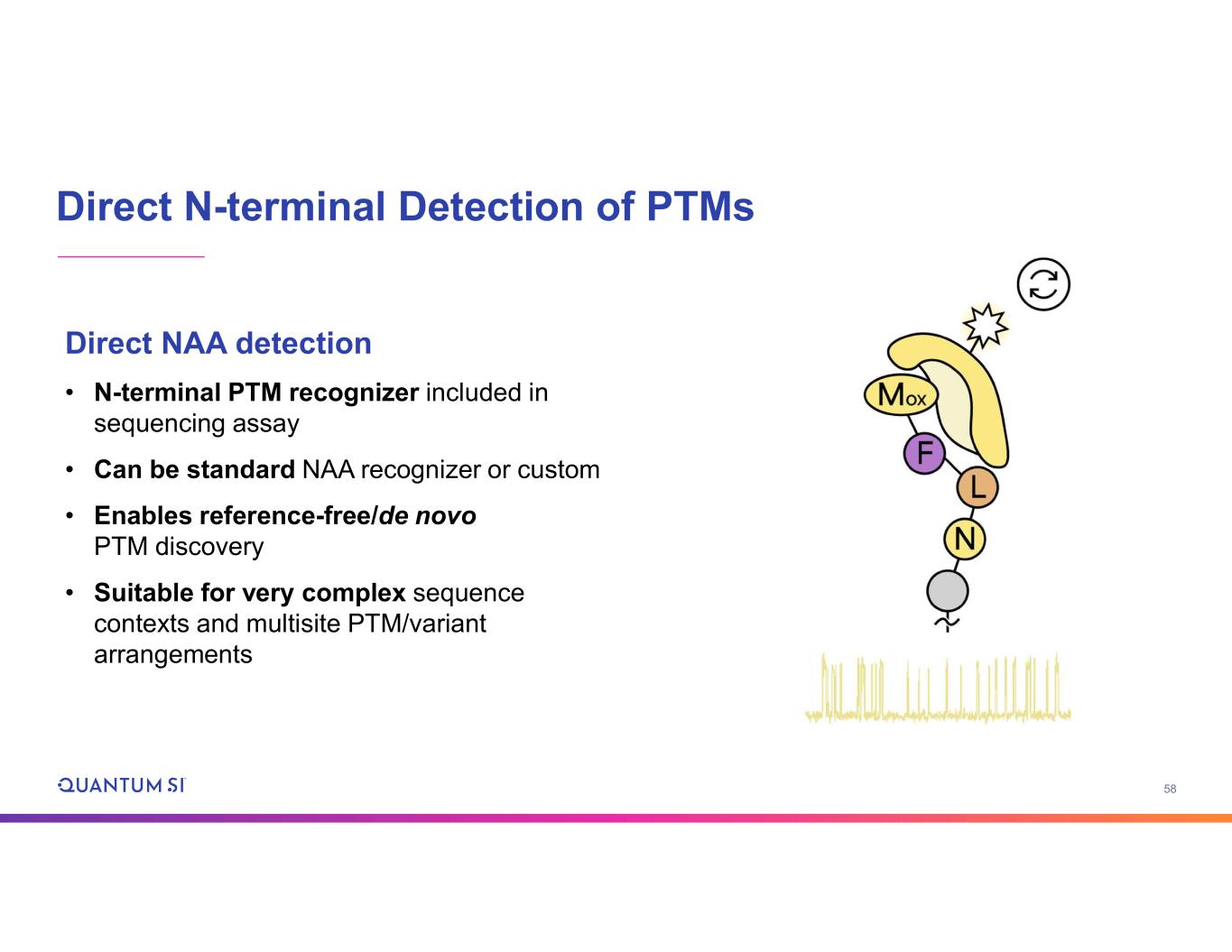
58 Direct N-terminal Detection of PTMs Direct NAA detection • N-terminal PTM recognizer included in sequencing assay • Can be standard NAA recognizer or custom • Enables reference-free/de novo PTM discovery • Suitable for very complex sequence contexts and multisite PTM/variant arrangements
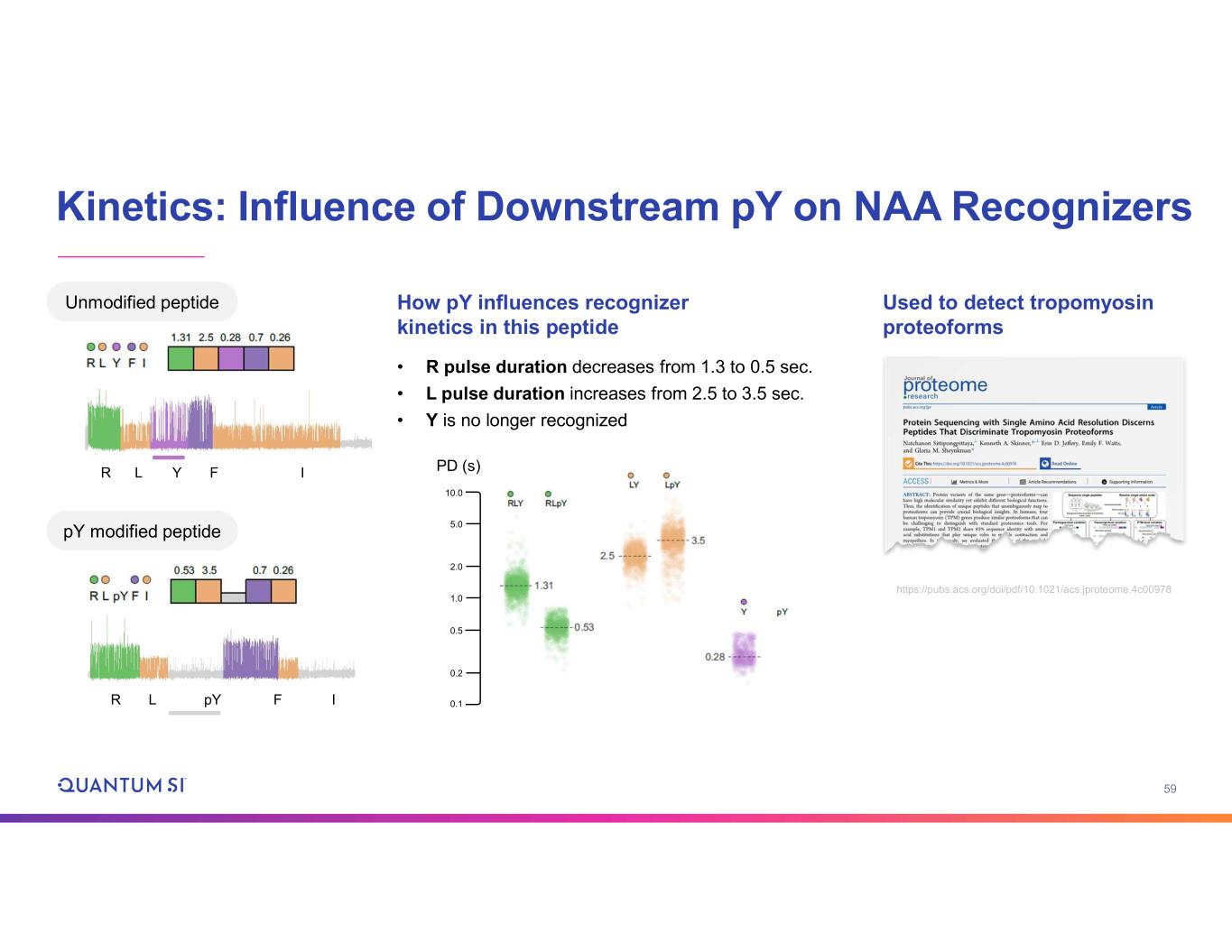
59 Kinetics: Influence of Downstream pY on NAA Recognizers Unmodified peptide • R pulse duration decreases from 1.3 to 0.5 sec. • L pulse duration increases from 2.5 to 3.5 sec. • Y is no longer recognized Used to detect tropomyosin proteoforms pY modified peptide R L pY F I R L Y F I https://pubs.acs.org/doi/pdf/10.1021/acs.jproteome.4c00978 How pY influences recognizer kinetics in this peptide 10.0 5.0 2.0 1.0 0.5 0.2 0.1 PD (s)
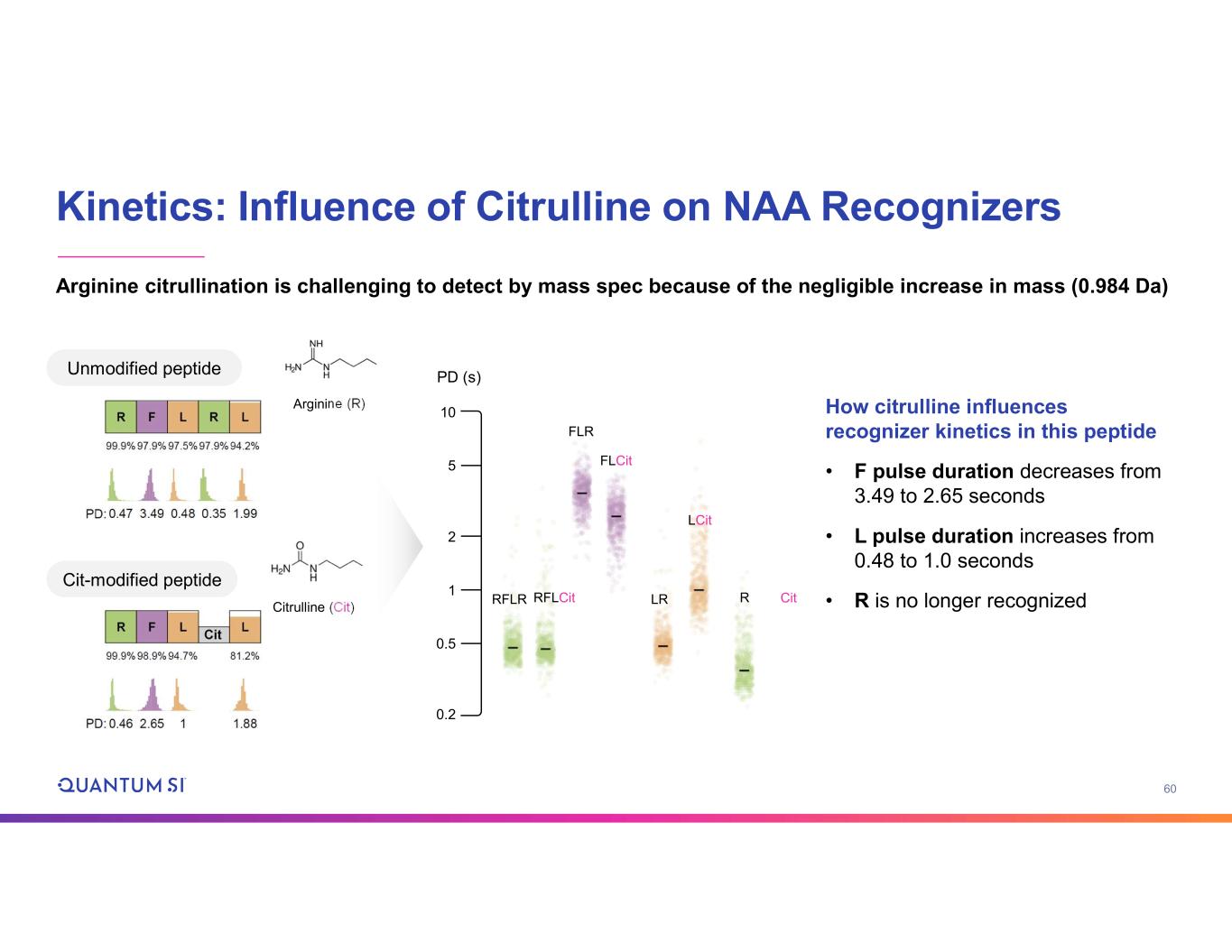
60 Kinetics: Influence of Citrulline on NAA Recognizers RFLR RFLCit FLCit FLR LCit LR R Cit Arginine (R) Citrulline (Cit) Unmodified peptide Cit-modified peptide How citrulline influences recognizer kinetics in this peptide • F pulse duration decreases from 3.49 to 2.65 seconds • L pulse duration increases from 0.48 to 1.0 seconds • R is no longer recognized Arginine citrullination is challenging to detect by mass spec because of the negligible increase in mass (0.984 Da) 10 5 2 1 0.5 0.2 PD (s)
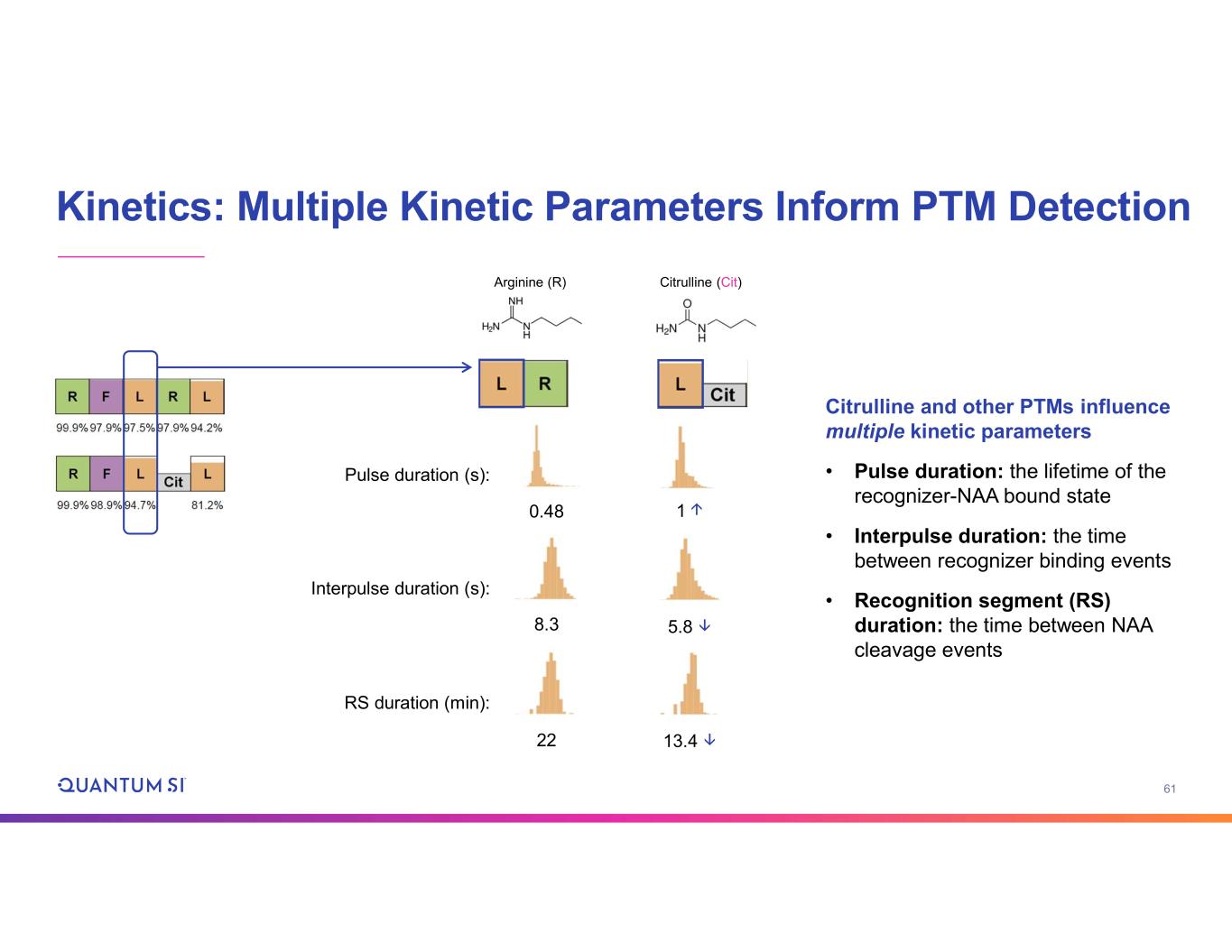
61 Kinetics: Multiple Kinetic Parameters Inform PTM Detection Arginine (R) Citrulline (Cit) Citrulline and other PTMs influence multiple kinetic parameters • Pulse duration: the lifetime of the recognizer-NAA bound state • Interpulse duration: the time between recognizer binding events • Recognition segment (RS) duration: the time between NAA cleavage events Pulse duration (s): 0.48 1↑ Interpulse duration (s): 8.3 5.8 ↓ RS duration (min): 22 13.4 ↓
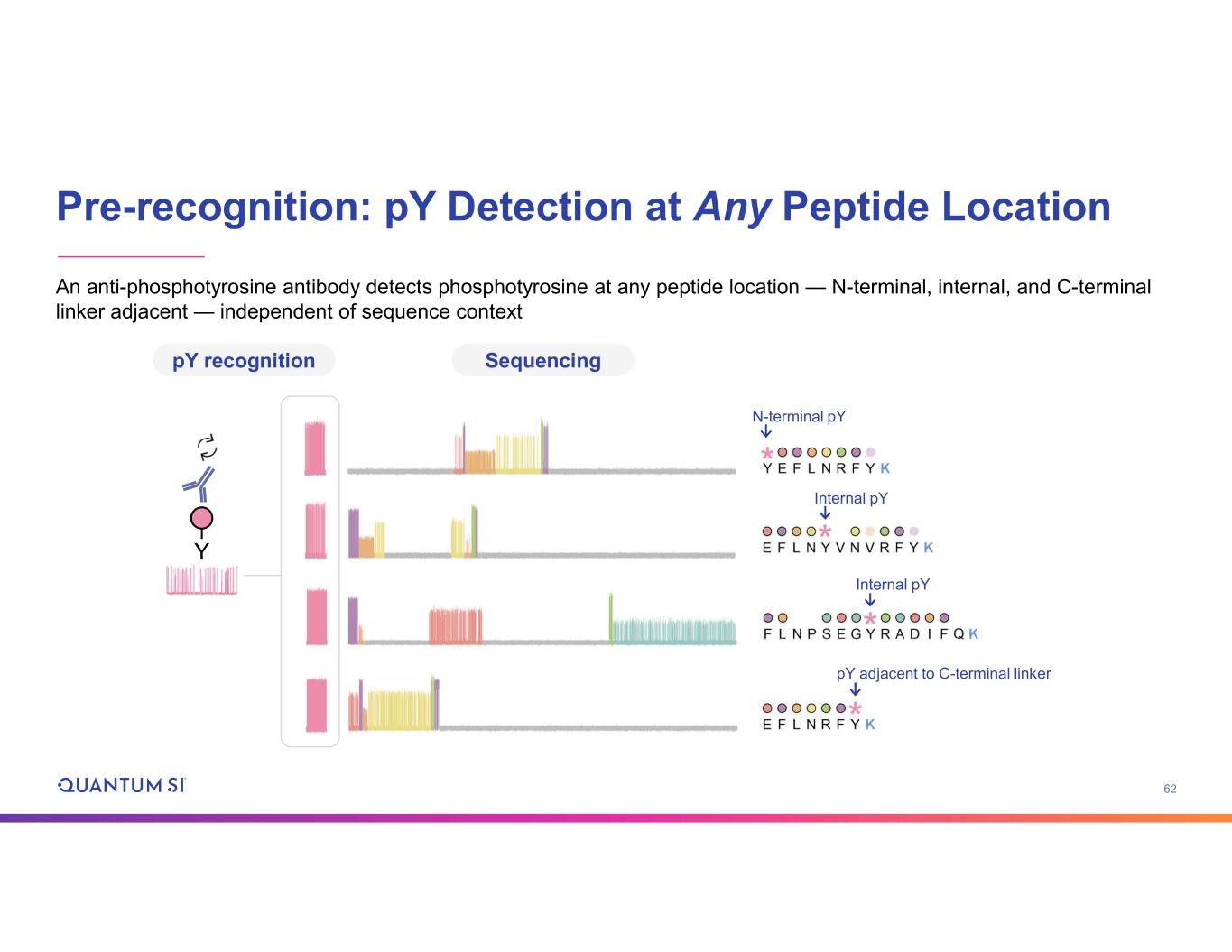
62 Pre-recognition: pY Detection at Any Peptide Location An anti-phosphotyrosine antibody detects phosphotyrosine at any peptide location — N-terminal, internal, and C-terminal linker adjacent — independent of sequence context pY recognition Sequencing Y N-terminal pY Internal pY Internal pY pY adjacent to C-terminal linker
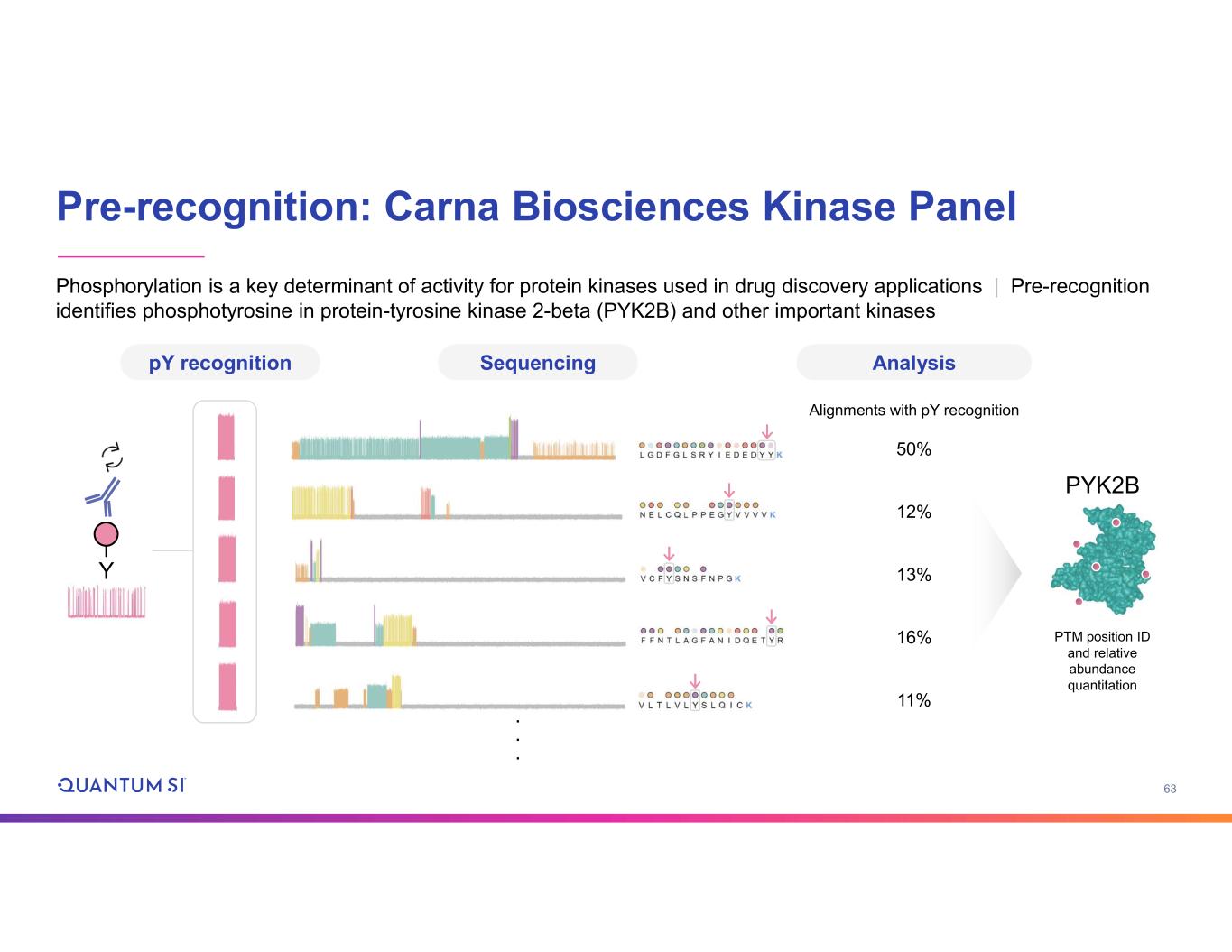
63 Pre-recognition: Carna Biosciences Kinase Panel pY recognition Sequencing Analysis 50% 12% 13% 16% 11% Alignments with pY recognition . . . Y PYK2B PTM position ID and relative abundance quantitation Phosphorylation is a key determinant of activity for protein kinases used in drug discovery applications | Pre-recognition identifies phosphotyrosine in protein-tyrosine kinase 2-beta (PYK2B) and other important kinases
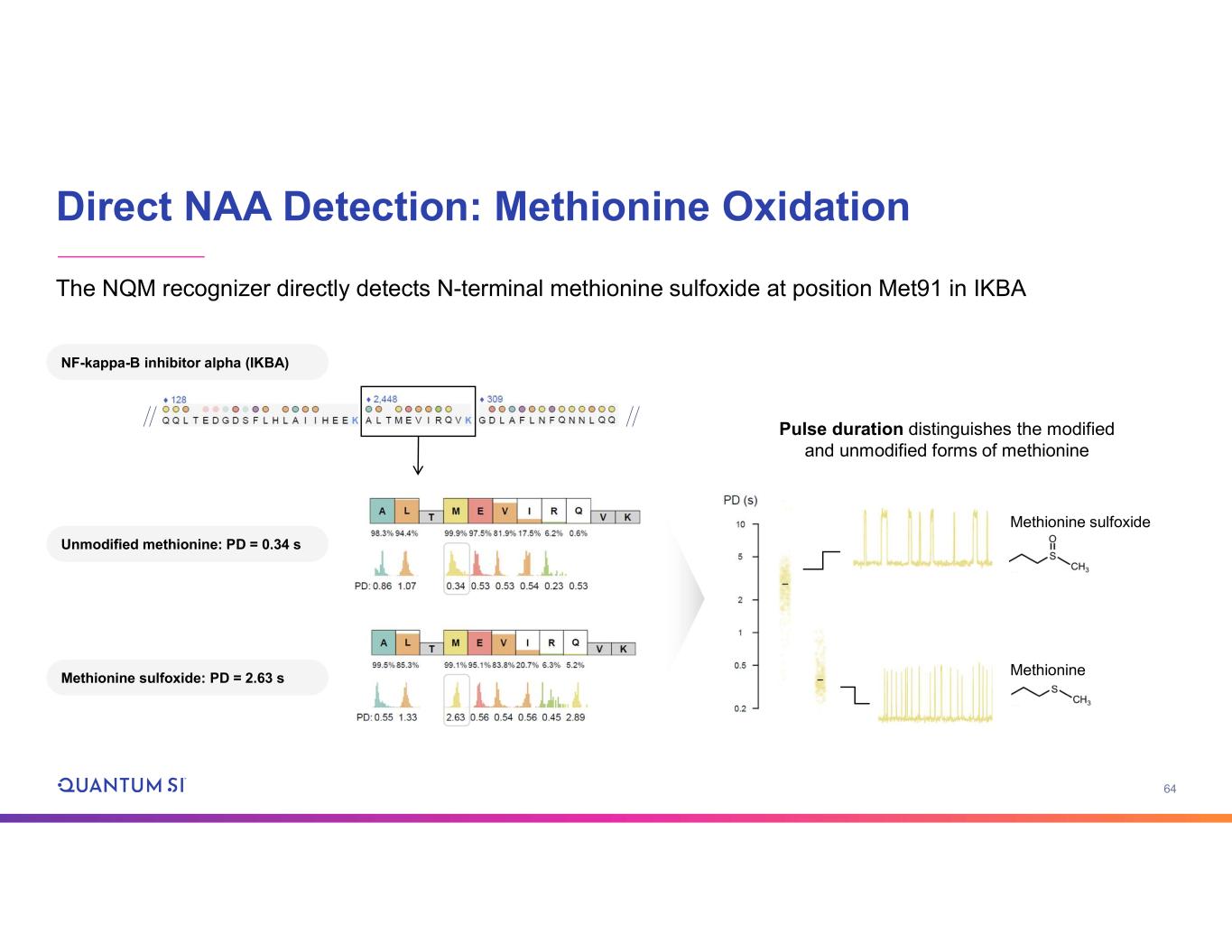
64 Direct NAA Detection: Methionine Oxidation Methionine sulfoxide Methionine Pulse duration distinguishes the modified and unmodified forms of methionine The NQM recognizer directly detects N-terminal methionine sulfoxide at position Met91 in IKBA NF-kappa-B inhibitor alpha (IKBA) Unmodified methionine: PD = 0.34 s Methionine sulfoxide: PD = 2.63 s
65 We Have a Clear Path to Broad PTM Coverage on Proteus Recognizer development Recognizers for all 20 amino acids will ensure maximum protein coverage and kinetic information available for PTM detection with all three methods Higher output on Proteus Enables deeper and more sensitive PTM detection and multiplexed panel- based solutions Artificial intelligence For data analysis to fully extract the massive volume of kinetic information in our NGPS output for PTM and variant detection and quantitation PTM access with three methods Three complimentary approaches that can be used in combination ensure access to the broadest range of PTMs anywhere in the proteome

66 The Road to Proteus Launch
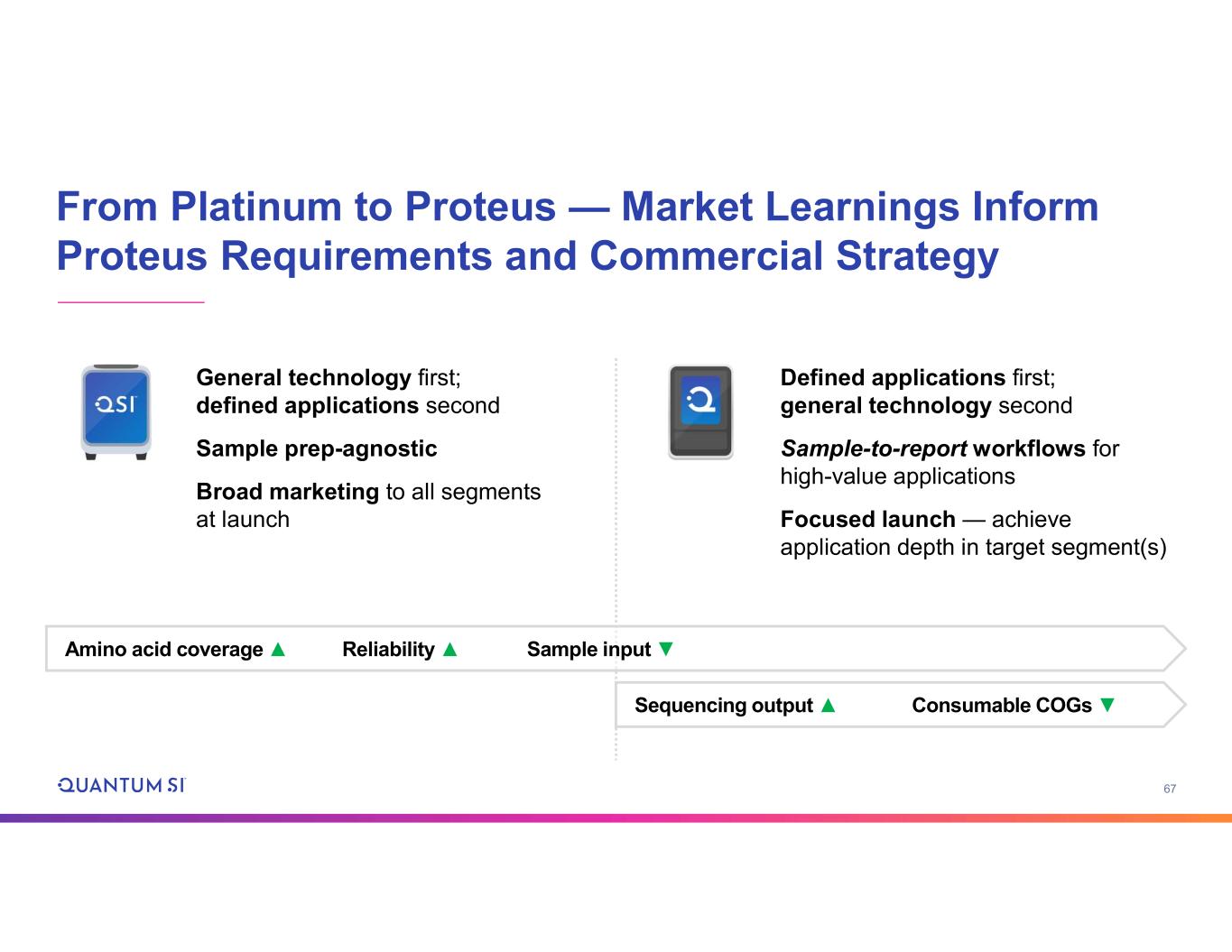
67 From Platinum to Proteus — Market Learnings Inform Proteus Requirements and Commercial Strategy General technology first; defined applications second Sample prep-agnostic Broad marketing to all segments at launch Defined applications first; general technology second Sample-to-report workflows for high-value applications Focused launch — achieve application depth in target segment(s) Amino acid coverage ▲ Reliability ▲ Sample input ▼ Sequencing output ▲ Consumable COGs ▼
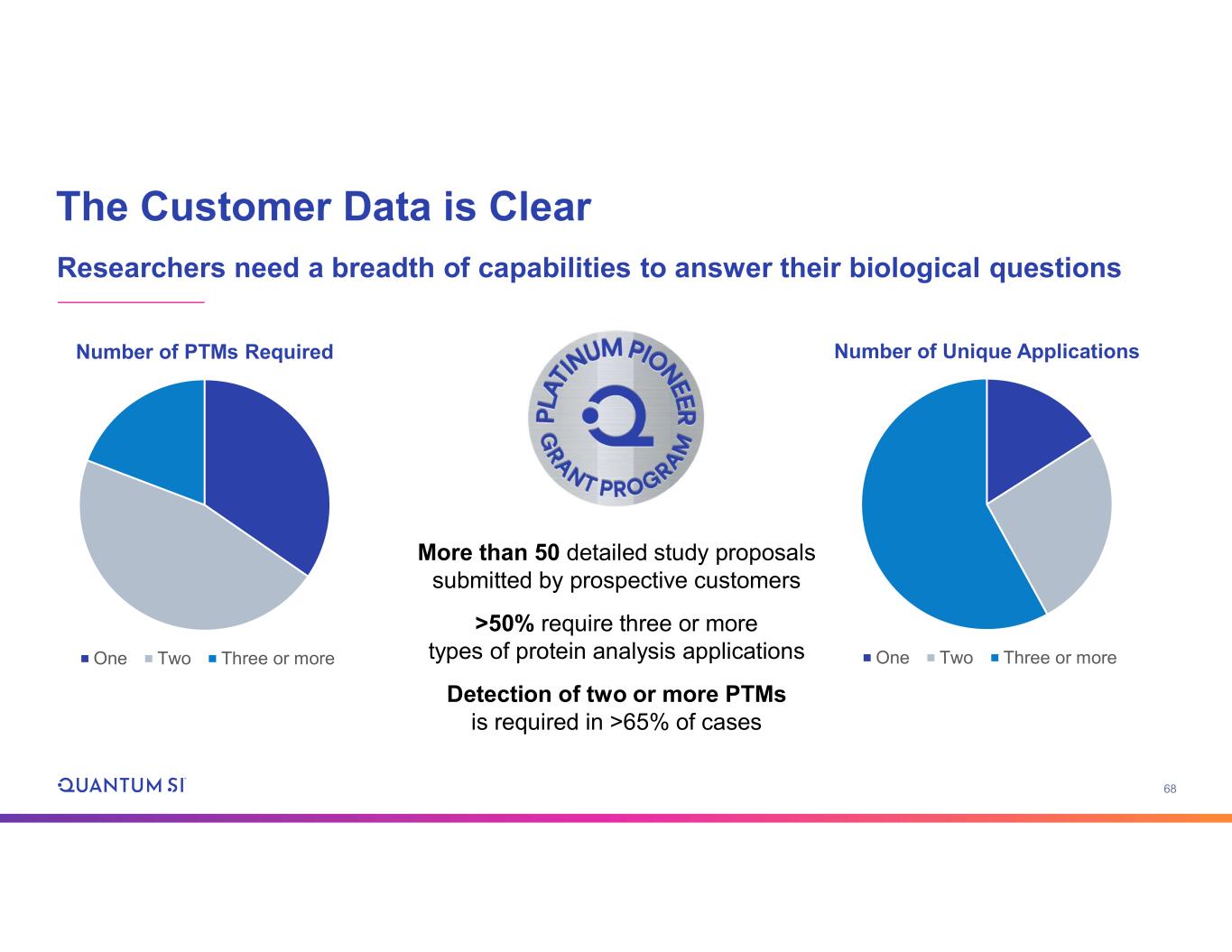
68 The Customer Data is Clear Researchers need a breadth of capabilities to answer their biological questions Number of Unique Applications One Two Three or more Number of PTMs Required One Two Three or more More than 50 detailed study proposals submitted by prospective customers >50% require three or more types of protein analysis applications Detection of two or more PTMs is required in >65% of cases

69 A Look Inside an Academic Medical Center Basic Research Lab Core Lab Translational Lab Clinical Lab Academic Medical Center • Wide range of application interests • Limited infrastructure • Lower utilization • Many technologies in use in one lab • Significant infrastructure • High utilization potential • Targeted applications; many involve PTMs • Value automation • High utilization potential • Defined application content; sample-to- report workflows • Automation is key • High utilization potential given repeat sample testing
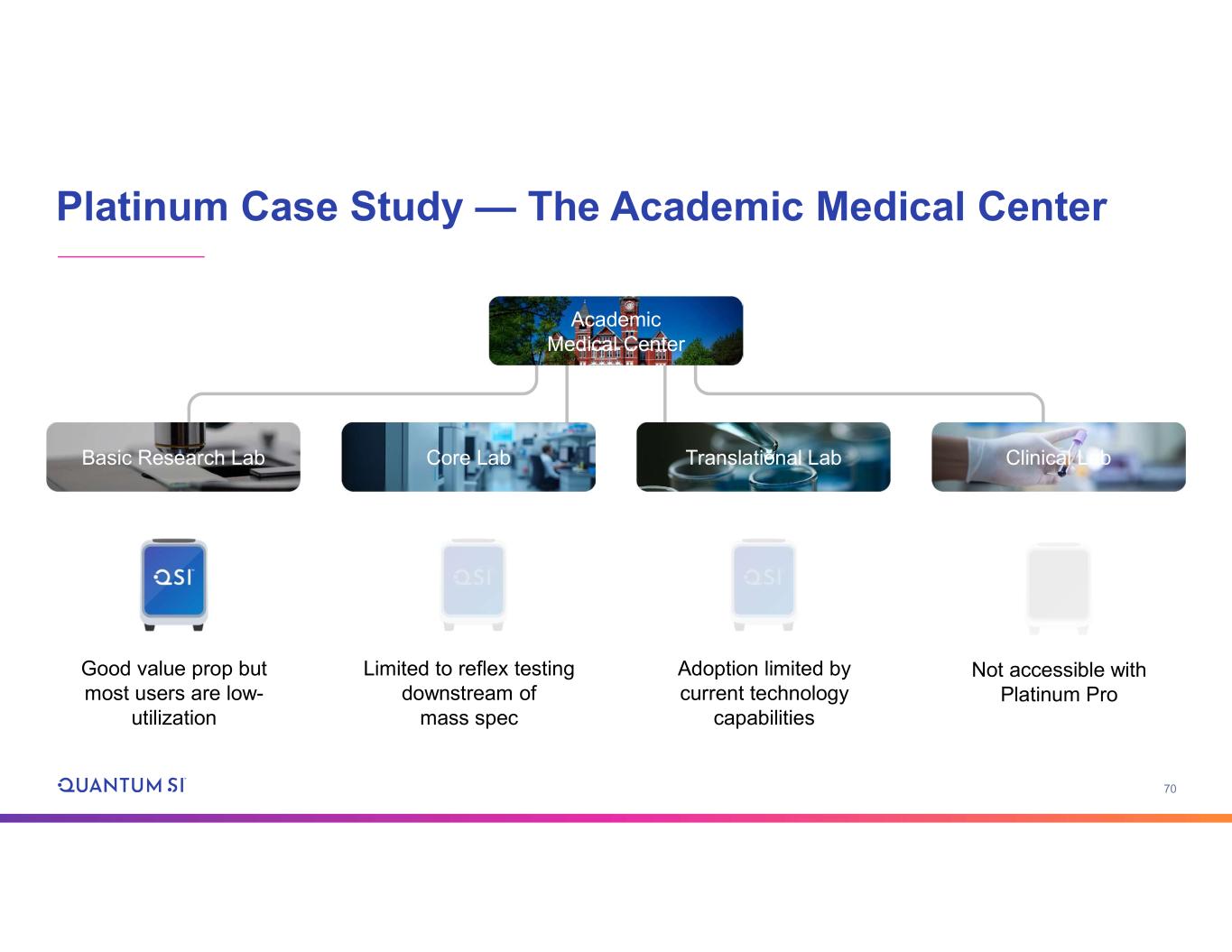
70 Platinum Case Study — The Academic Medical Center Basic Research Lab Core Lab Translational Lab Clinical Lab Academic Medical Center Good value prop but most users are low- utilization Limited to reflex testing downstream of mass spec Adoption limited by current technology capabilities Not accessible with Platinum Pro
71 The Increased Sequencing Output and Lower Costs of the Nanowell Array is Broadly Enabling More samples per run with 40× the output per consumable compared to Platinum Pro Deep analysis of a single sample to address complex mixtures or rare events like variants or PTMs Multiplexing samples to achieve higher throughput and lower costs per sample for cost sensitive applications like protein ID
72 Accelerating the Core Sequencing Technology and Analytics to Unlock the Highest-value Applications Robust PTM detection and multi-PTM profiling Single-amino acid variant detection Sequencing of variable regions of antibodies Biothreat detection and surveillance
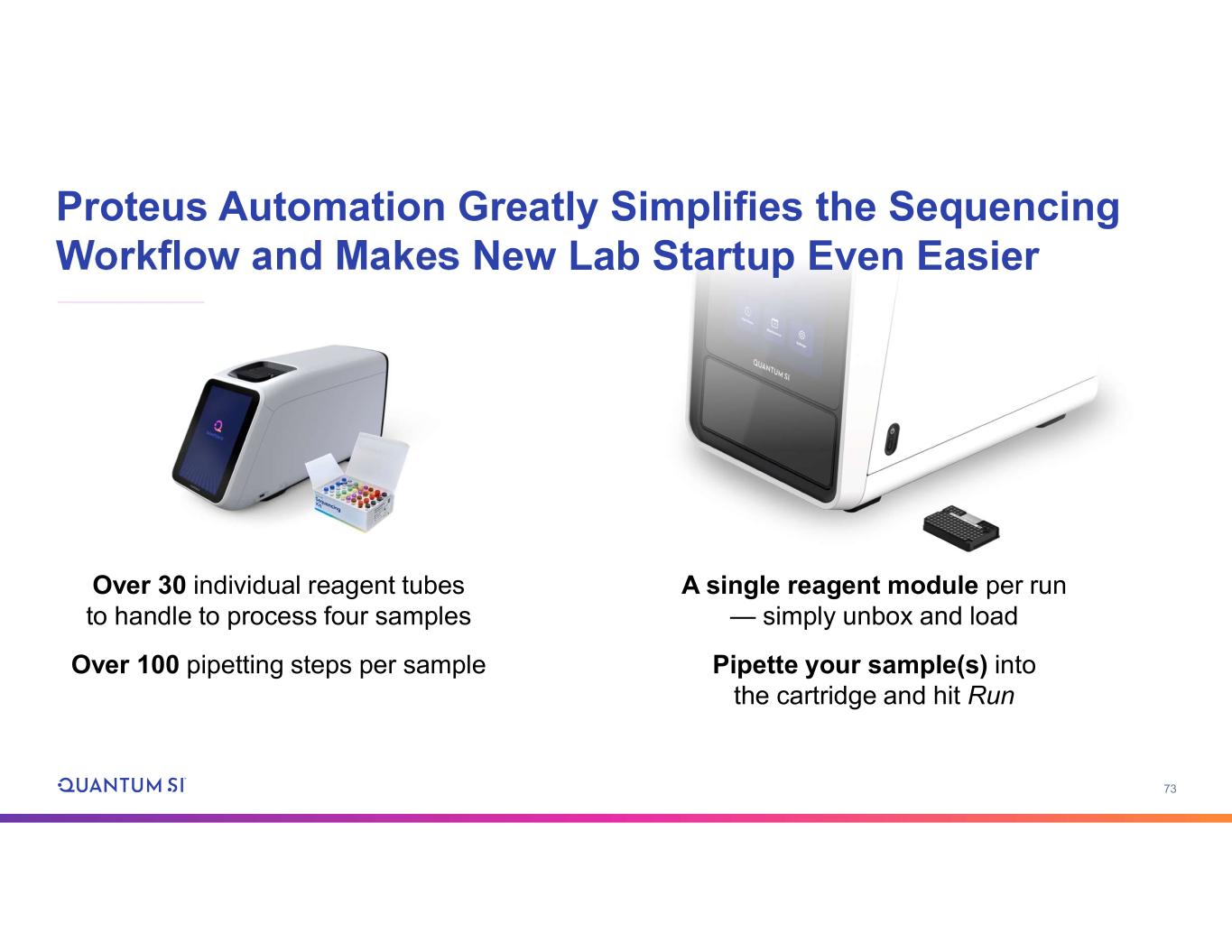
73 Proteus Automation Greatly Simplifies the Sequencing Workflow and Makes New Lab Startup Even Easier A single reagent module per run — simply unbox and load Pipette your sample(s) into the cartridge and hit Run Over 30 individual reagent tubes to handle to process four samples Over 100 pipetting steps per sample
74 Building a Partner Ecosystem to Accelerate Application Development + Deliver Sample-to-report Workflows Sample Prep + Enrichment Application Development Applications of AI for Data Analysis

75 New Industry Partnerships in PTMs and Ultra-low- abundance Protein Panels • Carna Biosciences is a leading provider of assay-grade kinase proteins to biopharma for drug discovery and profiling • Carna Biosciences is evaluating the use of QSI technology to assess and validate phosphorylation profiles of kinases • Collaboration presents the potential to leverage the complementary strengths and global reach of both organizations • Siena Quant is a SISCAPA Holdings Company — experts in the field of sample enrichment for proteomics • Siena Quant is enabling the quantitation of ultra-low-abundance biomarkers that have been historically unreliable to measure • QSI and Siena Quant will collaborate to develop sample-to-report workflows for customers to accurately measure clinically relevant biomarker panels

76 Proteus Will Increase the Depth of Penetration in Academic Medical Centers Compared to Platinum Single-molecule protein sequencing will remain a useful tool for basic research High-value application capabilities + increased throughput = first-line platform Expanded PTM capabilities + high-value targeted panels + automation Automation + sample-to- report workflows opens this lab for Proteus Basic Research Lab Core Lab Translational Lab Clinical Lab Academic Medical Center

77 Proteus Will Enable Single-molecule Protein Sequencing Across a Broad Range of End Markets Academic Research Pharma + Biotech Industrial Defense Clinical Diagnostics Translational Research Agriculture
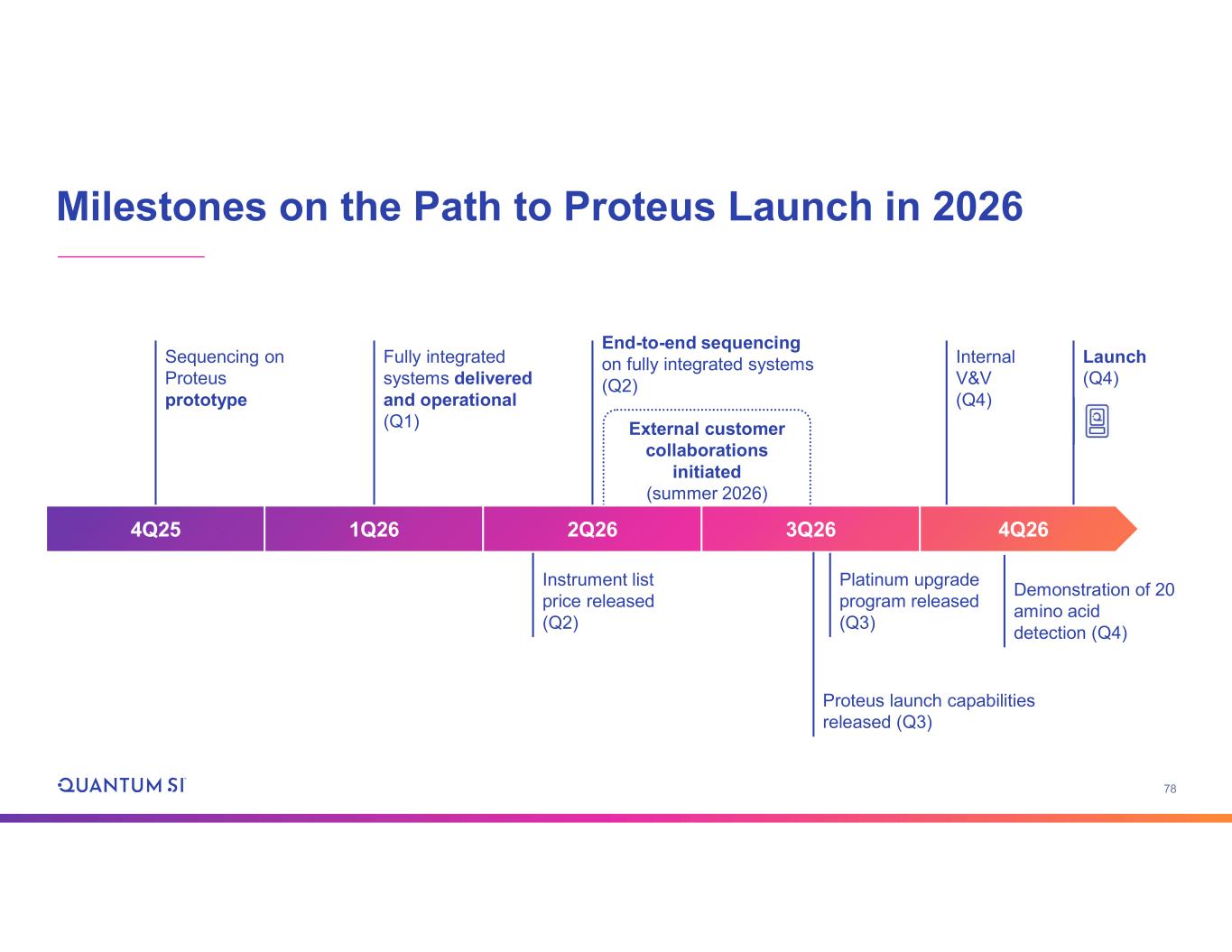
78 Milestones on the Path to Proteus Launch in 2026 External customer collaborations initiated (summer 2026) End-to-end sequencing on fully integrated systems (Q2) Platinum upgrade program released (Q3) Instrument list price released (Q2) Internal V&V (Q4) Sequencing on Proteus prototype Fully integrated systems delivered and operational (Q1) Launch (Q4) 4Q25 1Q26 2Q26 3Q26 4Q26 Demonstration of 20 amino acid detection (Q4) Proteus launch capabilities released (Q3)

79 QSI is Well-positioned to Build Upon Our First-to-market Advantage and Extend Our Leadership into the Future Extend our technology lead through best-in- class R&D innovation + execution Execute on development roadmap to deliver a compelling Proteus launch Build a partner ecosystem to accelerate application development + deliver sample-to-report workflows Grow user base with Platinum Pro and develop the market for Proteus launch