

October 2025 Company Presentation

Forward-Looking Statements This presentation contains forward-looking statements within the meaning of the Private Securities Litigation Reform Act of 1995, as amended. These statements may be identified by words such as “aims,” “anticipates,” “believes,” “could,” “estimates,” “expects,” “forecasts,” “goal,” “intends,” “may,” “plans,” “possible,” “potential,” “seeks,” “will,” and variations of these words or similar expressions that are intended to identify forward-looking statements. Any such statements in this presentation that are not statements of historical fact may be deemed to be forward-looking statements. These forward-looking statements include, without limitation, statements regarding the potential of our EDO platform to deliver higher levels of oligonucleotide to the nuclei, the therapeutic potential and safety profile of PGN-EDODM1 based on data from the 5, 10 and 15 mg/kg cohorts of the FREEDOM-DM1 study, our expectations regarding the potential for significant correction of mis-splicing with more doses of PGN-EDODM1 over a longer treatment period to potentially provide improved functional benefit for patients with DM1, the design, initiation and conduct of clinical trials, including expected timelines for additional data reports from our FREEDOM-DM1 trial and the initial data report from our FREEDOM2-DM1 trial, the potential for any functional improvements that may result from robust splicing correction with PGN-EDODM1, dose-dependent increases in splicing suggesting that PGN-EDODM1 is getting into the muscle and effectively binding to the target, ongoing and planned regulatory interactions and our financial resources and expected cash runway. Any forward-looking statements in this presentation are based on current expectations, estimates and projections only as of the date of this presentation and are subject to a number of risks and uncertainties that could cause actual results to differ materially and adversely from those set forth in or implied by such forward-looking statements. These risks and uncertainties include, but are not limited to: delays or failure to successfully initiate or complete our ongoing and planned development activities for our product candidates, including PGN-EDODM1; our ability to enroll patients in our clinical trials, including FREEDOM2, that our interpretation of clinical and preclinical study results may be incorrect, or that we may not observe the levels of therapeutic activity in clinical testing that we anticipate based on prior clinical or preclinical results, including for PGN-EDODM1; our product candidates, including PGN-EDODM1, may not be safe and effective or otherwise demonstrate safety and efficacy in our clinical trials; adverse outcomes from our regulatory interactions, including delays in regulatory review, clearance to proceed or approval by regulatory authorities with respect to our programs, including clearance to commence planned clinical studies of our product candidates, or other regulatory feedback requiring modifications to our development programs, including in each case with respect to our including FREEDOM and FREEDOM2 clinical trials; changes in regulatory framework that are out of our control; our ability to obtain, maintain and protect our intellectual property; our ability to enforce our patents against infringers and defend our patent portfolio against challenges from third parties; competition from others developing therapies for the indications we are pursuing; unexpected increases in the expenses associated with our development activities or other events that adversely impact our financial resources and cash runway; and our dependence on third parties for some or all aspects of our product manufacturing, research and preclinical and clinical testing. Additional risks concerning PepGen's programs and operations are described in our most recent filings with the SEC. PepGen explicitly disclaims any obligation to update any forward-looking statements except to the extent required by law. This presentation discusses PGN-EDODM1, an investigational therapy, that has not been approved for use in any country, and is not intended to convey conclusions about their efficacy or safety. There is no guarantee that PGN-EDODM1 or any other investigational therapy will successfully complete clinical development or gain regulatory authority approval.
Leveraging EDO Platform to Drive Meaningful Impact for Patients PGN-EDODM1: Myotonic Dystrophy Type 1 Best-in-class potential; selectively targets only pathogenic DMPK RNA Favorable emerging safety profile and highest splicing correction ever reported in patients Orphan Drug & Fast Track Designation (U.S.); Orphan Drug Designation (EU) Developing research pipeline that applies platform differentiators to address underlying neuromuscular disease drivers Exploring EDO's potential in genetic conditions, including Charcot-Marie-Tooth disease Research Pipeline EDO PLATFORM Achieving superior nuclear delivery and uptake of therapeutic oligonucleotides, overcoming key limitations of prior approaches Backed by a team of leading neuromuscular researchers with deep expertise in genetic disease biology and oligonucleotide drug development OUR VISION Develop therapies that address the root cause of serious genetic neuromuscular and neurological diseases—driving meaningful, functional improvement Q1 2026: FREEDOM2 5 mg/kg clinical results H2 2026: FREEDOM2 10 mg/kg clinical results Upcoming Milestones Cash runway into 2H 2027
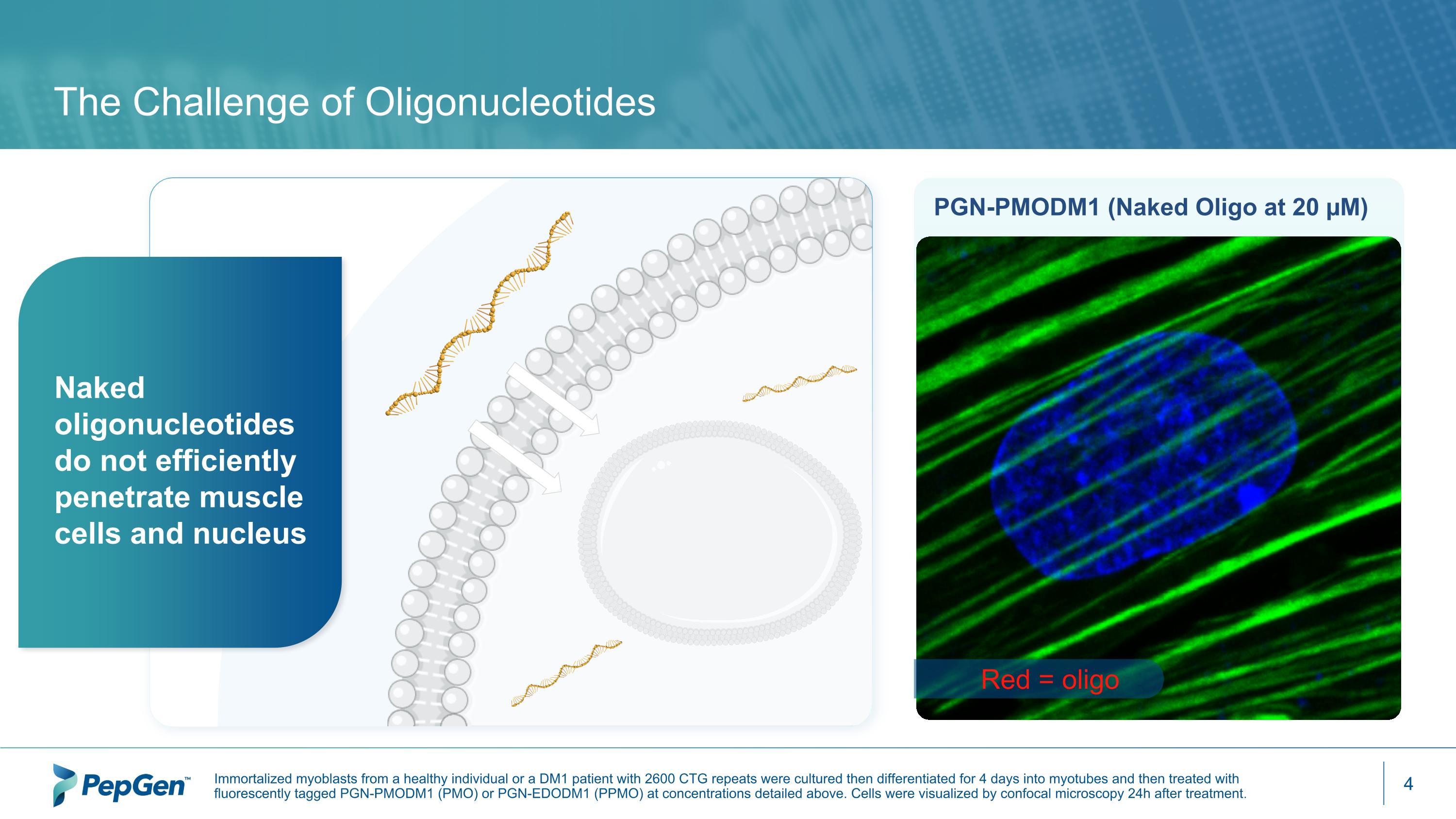
The Challenge of Oligonucleotides PGN-PMODM1 (Naked Oligo at 20 µM) Red = oligo Naked oligonucleotides do not efficiently penetrate muscle cells and nucleus Immortalized myoblasts from a healthy individual or a DM1 patient with 2600 CTG repeats were cultured then differentiated for 4 days into myotubes and then treated with fluorescently tagged PGN-PMODM1 (PMO) or PGN-EDODM1 (PPMO) at concentrations detailed above. Cells were visualized by confocal microscopy 24h after treatment.
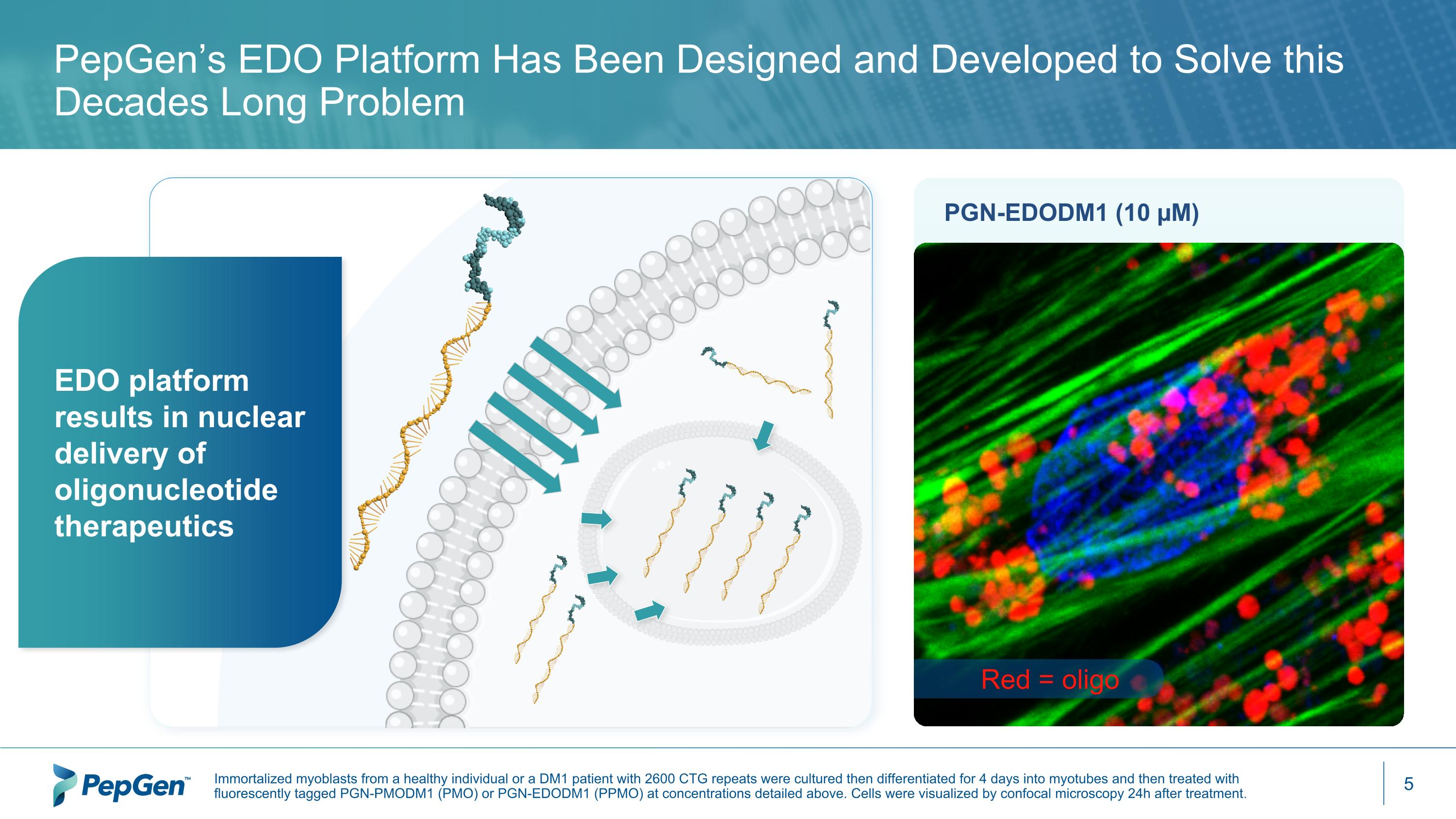
PGN-EDODM1 (10 µM) Red = oligo PepGen’s EDO Platform Has Been Designed and Developed to Solve this Decades Long Problem EDO platform results in nuclear delivery of oligonucleotide therapeutics Immortalized myoblasts from a healthy individual or a DM1 patient with 2600 CTG repeats were cultured then differentiated for 4 days into myotubes and then treated with fluorescently tagged PGN-PMODM1 (PMO) or PGN-EDODM1 (PPMO) at concentrations detailed above. Cells were visualized by confocal microscopy 24h after treatment.
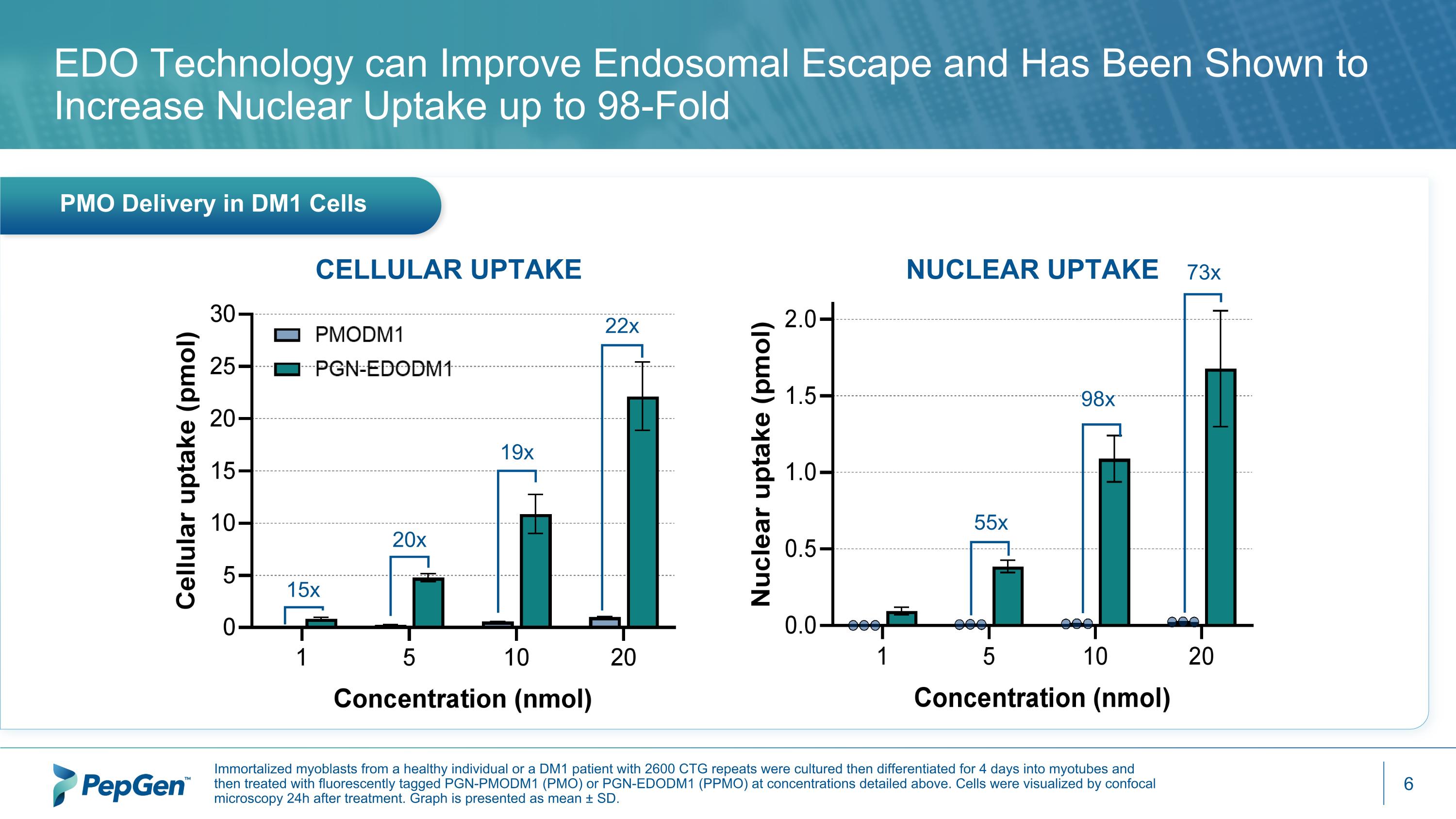
15x 20x 19x 22x 55x 98x 73x PMO Delivery in DM1 Cells EDO Technology can Improve Endosomal Escape and Has Been Shown to Increase Nuclear Uptake up to 98-Fold CELLULAR UPTAKE NUCLEAR UPTAKE Immortalized myoblasts from a healthy individual or a DM1 patient with 2600 CTG repeats were cultured then differentiated for 4 days into myotubes and then treated with fluorescently tagged PGN-PMODM1 (PMO) or PGN-EDODM1 (PPMO) at concentrations detailed above. Cells were visualized by confocal microscopy 24h after treatment. Graph is presented as mean ± SD.

PGN-EDODM1 – Myotonic Dystrophy Type 1 (DM1)
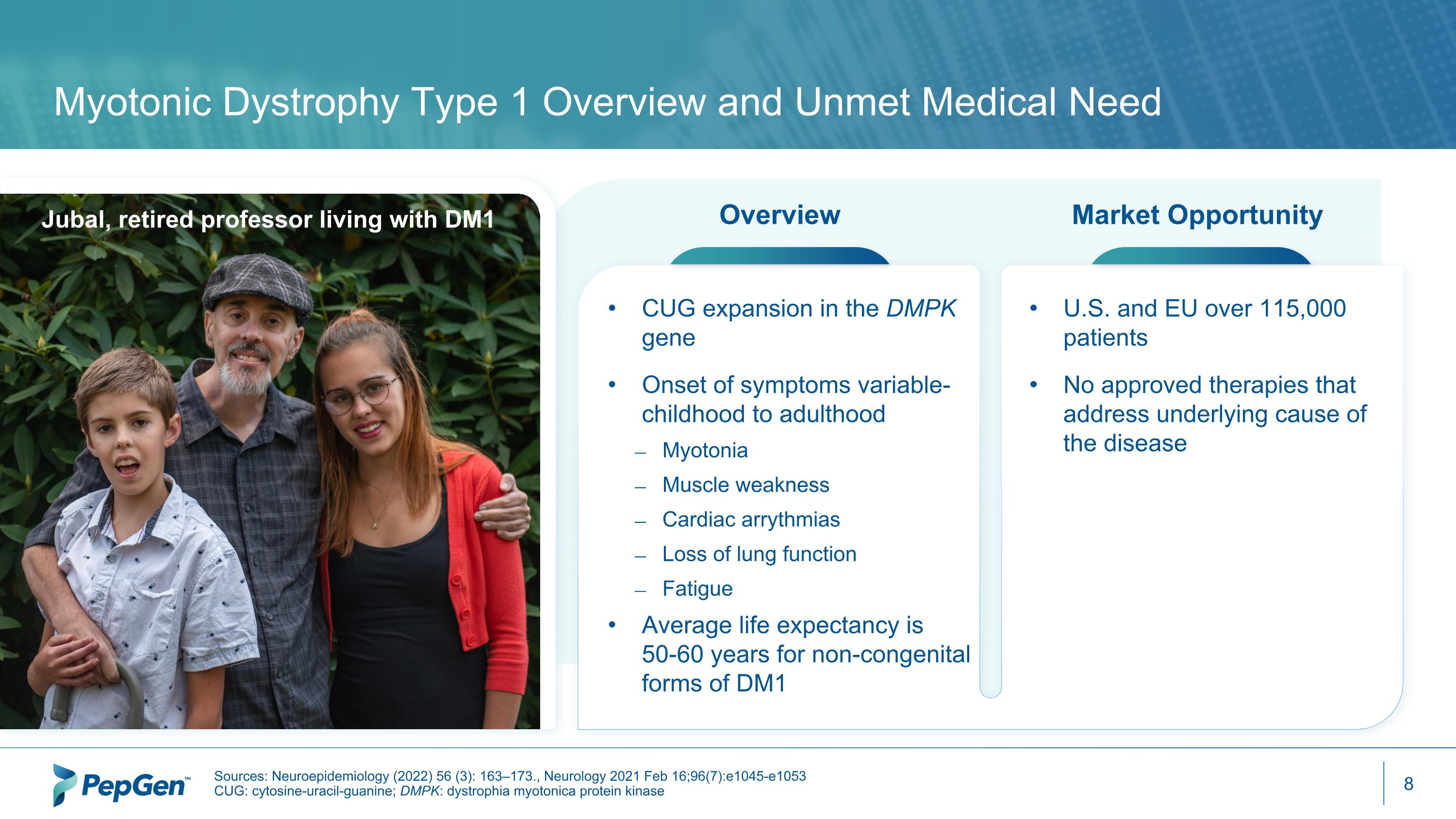
Myotonic Dystrophy Type 1 Overview and Unmet Medical Need Sources: Neuroepidemiology (2022) 56 (3): 163–173., Neurology 2021 Feb 16;96(7):e1045-e1053 CUG: cytosine-uracil-guanine; DMPK: dystrophia myotonica protein kinase Overview Market Opportunity CUG expansion in the DMPK gene Onset of symptoms variable- childhood to adulthood Myotonia Muscle weakness Cardiac arrythmias Loss of lung function Fatigue Average life expectancy is 50-60 years for non-congenital forms of DM1 U.S. and EU over 115,000 patients No approved therapies that address underlying cause of the disease Jubal, retired professor living with DM1
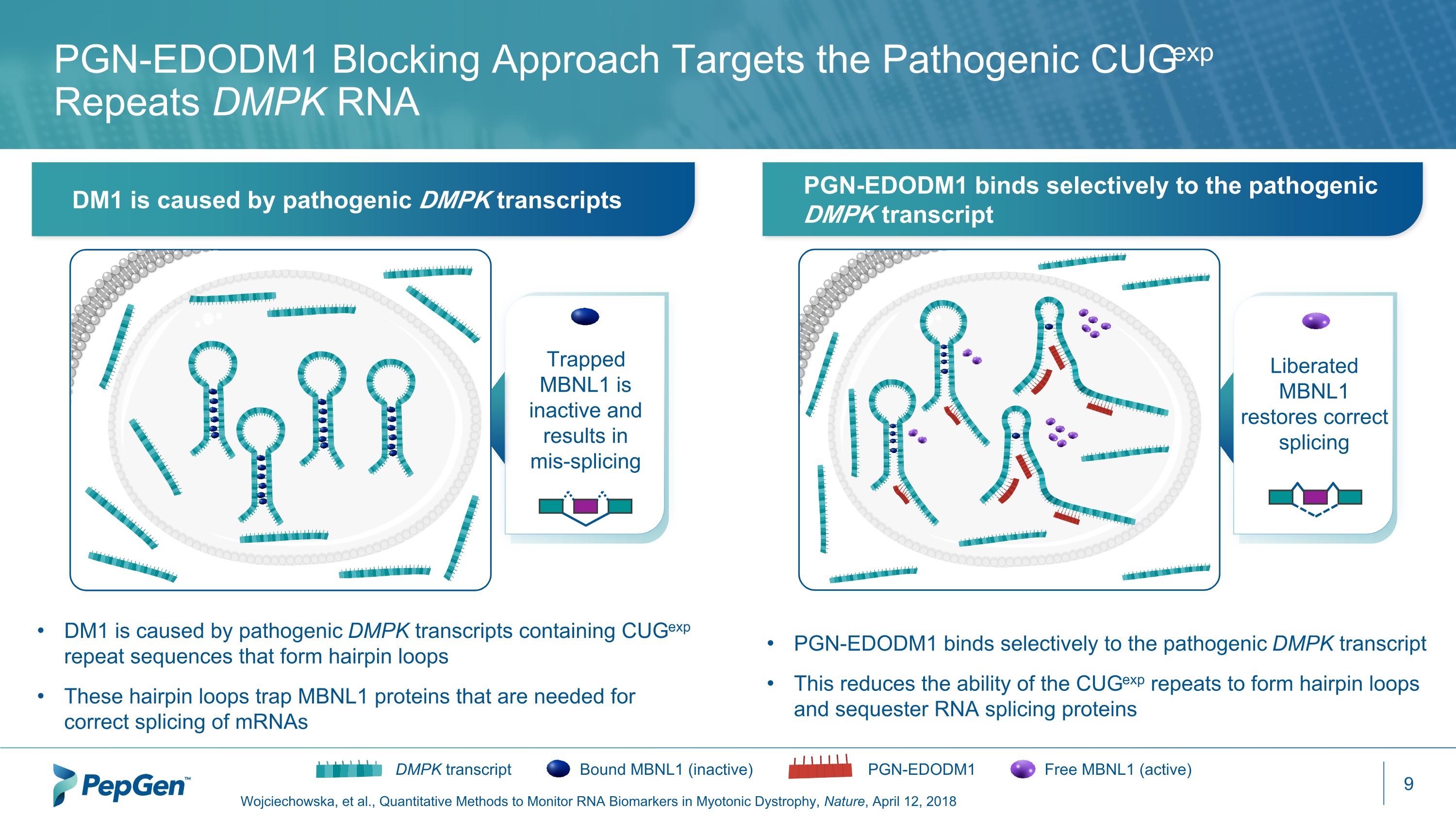
PGN-EDODM1 Blocking Approach Targets the Pathogenic CUGexp Repeats DMPK RNA Wojciechowska, et al., Quantitative Methods to Monitor RNA Biomarkers in Myotonic Dystrophy, Nature, April 12, 2018 DM1 is caused by pathogenic DMPK transcripts PGN-EDODM1 binds selectively to the pathogenic DMPK transcript Bound MBNL1 (inactive) DMPK transcript Free MBNL1 (active) PGN-EDODM1 PGN-EDODM1 binds selectively to the pathogenic DMPK transcript This reduces the ability of the CUGexp repeats to form hairpin loops and sequester RNA splicing proteins DM1 is caused by pathogenic DMPK transcripts containing CUGexp repeat sequences that form hairpin loops These hairpin loops trap MBNL1 proteins that are needed for correct splicing of mRNAs Trapped MBNL1 is inactive and results in mis-splicing Liberated MBNL1 restores correct splicing
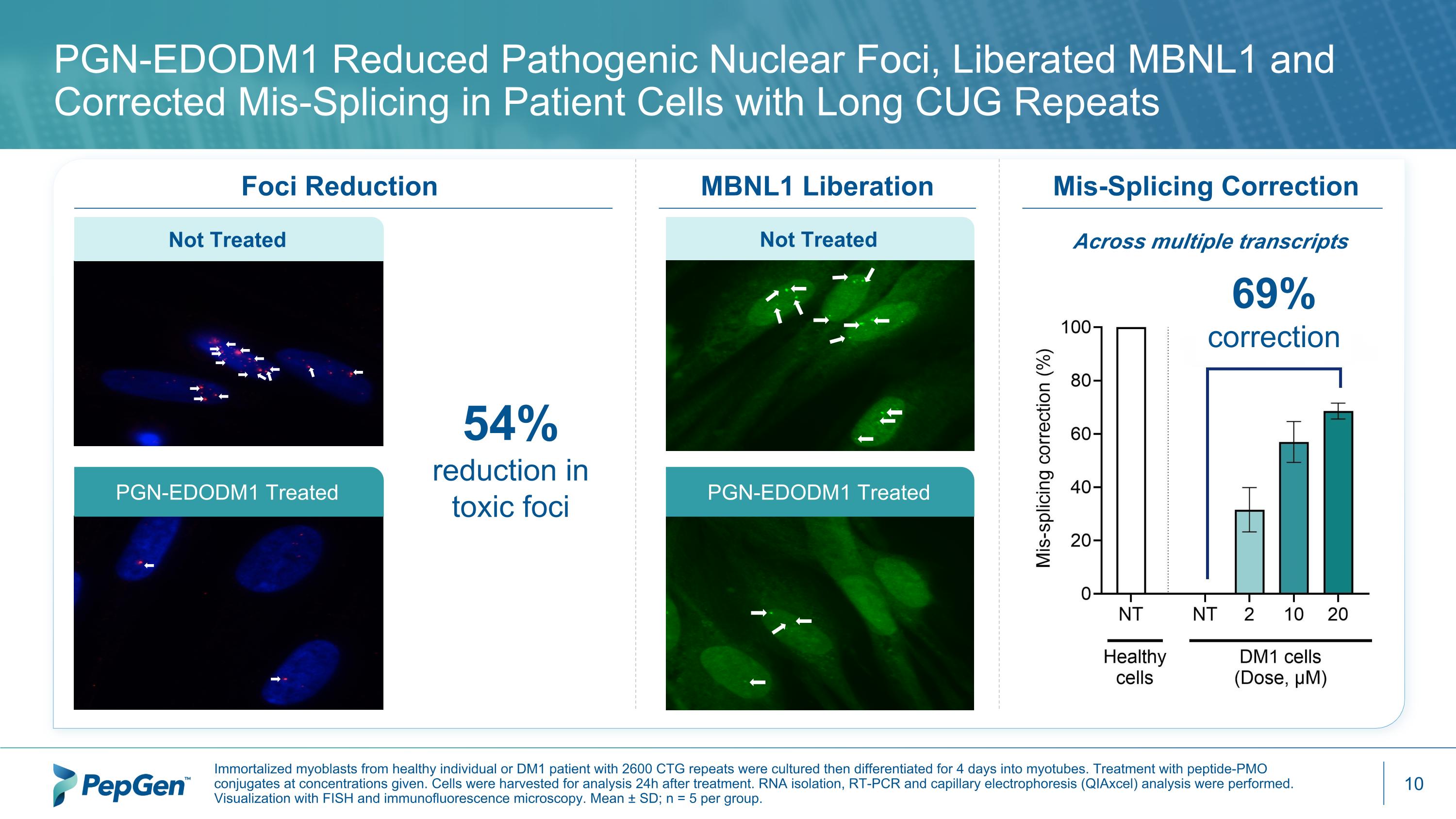
MBNL1 Liberation Foci Reduction PGN-EDODM1 Reduced Pathogenic Nuclear Foci, Liberated MBNL1 and Corrected Mis-Splicing in Patient Cells with Long CUG Repeats Immortalized myoblasts from healthy individual or DM1 patient with 2600 CTG repeats were cultured then differentiated for 4 days into myotubes. Treatment with peptide-PMO conjugates at concentrations given. Cells were harvested for analysis 24h after treatment. RNA isolation, RT-PCR and capillary electrophoresis (QIAxcel) analysis were performed. Visualization with FISH and immunofluorescence microscopy. Mean ± SD; n = 5 per group. Not Treated PGN-EDODM1 Treated Not Treated PGN-EDODM1 Treated 54% reduction in toxic foci Across multiple transcripts Mis-Splicing Correction 69% correction
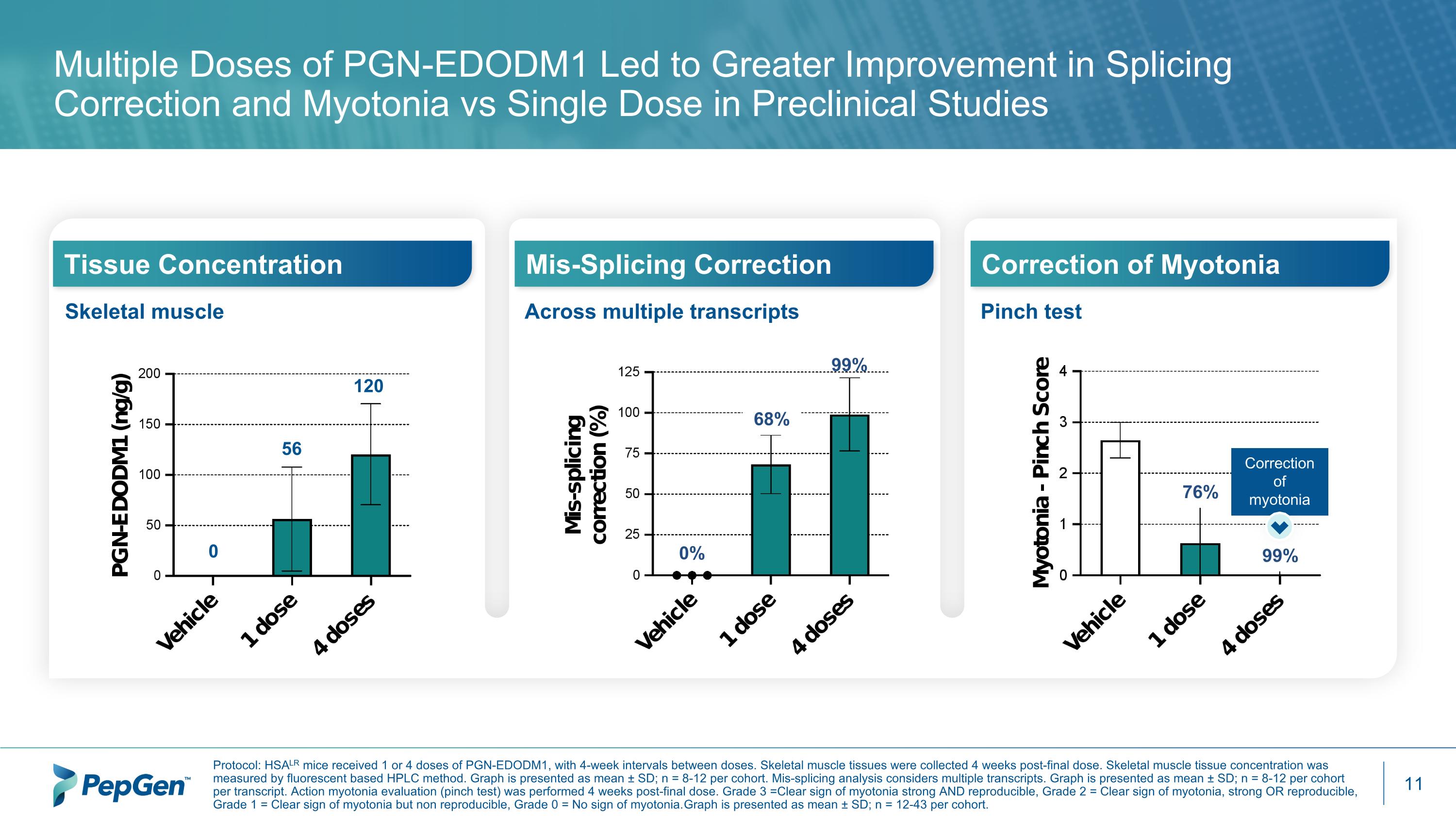
Multiple Doses of PGN-EDODM1 Led to Greater Improvement in Splicing Correction and Myotonia vs Single Dose in Preclinical Studies Protocol: HSALR mice received 1 or 4 doses of PGN-EDODM1, with 4-week intervals between doses. Skeletal muscle tissues were collected 4 weeks post-final dose. Skeletal muscle tissue concentration was measured by fluorescent based HPLC method. Graph is presented as mean ± SD; n = 8-12 per cohort. Mis-splicing analysis considers multiple transcripts. Graph is presented as mean ± SD; n = 8-12 per cohort per transcript. Action myotonia evaluation (pinch test) was performed 4 weeks post-final dose. Grade 3 = Clear sign of myotonia strong AND reproducible, Grade 2 = Clear sign of myotonia, strong OR reproducible, Grade 1 = Clear sign of myotonia but non reproducible, Grade 0 = No sign of myotonia. Graph is presented as mean ± SD; n = 12-43 per cohort. Skeletal muscle Tissue Concentration Across multiple transcripts Mis-Splicing Correction Correction of Myotonia Pinch test 56 120 0 68% 99% 0% 76% 99% Correction of myotonia
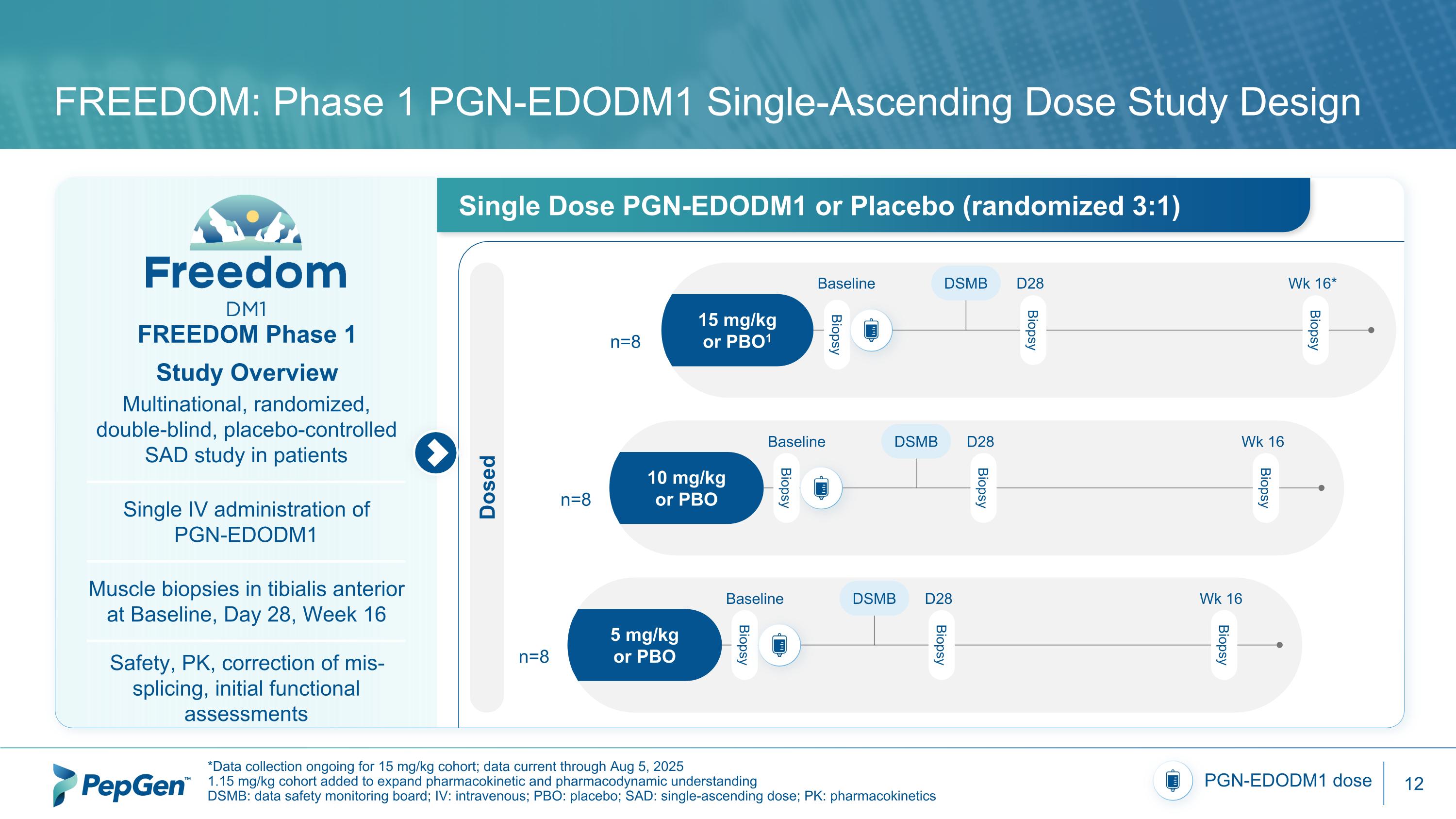
FREEDOM: Phase 1 PGN-EDODM1 Single-Ascending Dose Study Design *Data collection ongoing for 15 mg/kg cohort; data current through Aug 5, 2025 1.15 mg/kg cohort added to expand pharmacokinetic and pharmacodynamic understanding DSMB: data safety monitoring board; IV: intravenous; PBO: placebo; SAD: single-ascending dose; PK: pharmacokinetics PGN-EDODM1 dose FREEDOM Phase 1 Study Overview Multinational, randomized, double-blind, placebo-controlled SAD study in patients Single IV administration of PGN-EDODM1 Muscle biopsies in tibialis anterior at Baseline, Day 28, Week 16 Safety, PK, correction of mis-splicing, initial functional assessments Single Dose PGN-EDODM1 or Placebo (randomized 3:1) 15 mg/kg or PBO1 Baseline DSMB Wk 16* D28 Biopsy Biopsy Biopsy 10 mg/kg or PBO Baseline DSMB Wk 16 D28 Biopsy Biopsy Biopsy Dosed n=8 n=8 5 mg/kg or PBO Baseline DSMB Wk 16 D28 Biopsy Biopsy Biopsy n=8
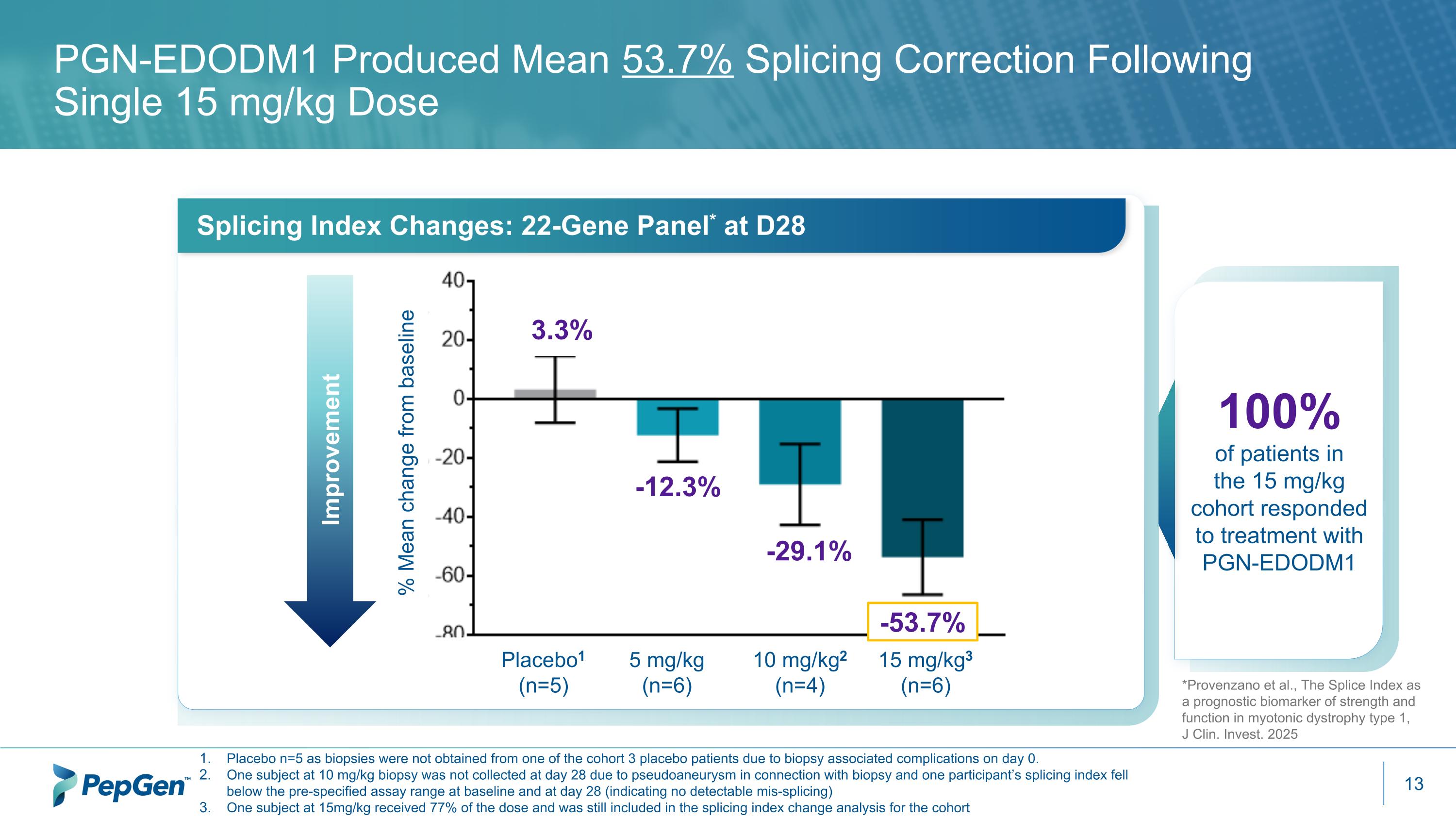
PGN-EDODM1 Produced Mean 53.7% Splicing Correction Following Single 15 mg/kg Dose Placebo n=5 as biopsies were not obtained from one of the cohort 3 placebo patients due to biopsy associated complications on day 0. One subject at 10 mg/kg biopsy was not collected at day 28 due to pseudoaneurysm in connection with biopsy and one participant’s splicing index fell below the pre-specified assay range at baseline and at day 28 (indicating no detectable mis-splicing) One subject at 15mg/kg received 77% of the dose and was still included in the splicing index change analysis for the cohort Splicing Index Changes: 22-Gene Panel* at D28 Improvement -53.7% -29.1% 3.3% -12.3% 100% of patients in the 15 mg/kg cohort responded to treatment with PGN-EDODM1 % Mean change from baseline Placebo1 (n=5) 5 mg/kg (n=6) 10 mg/kg2 (n=4) 15 mg/kg3 (n=6) *Provenzano et al., The Splice Index as a prognostic biomarker of strength and function in myotonic dystrophy type 1, J Clin. Invest. 2025
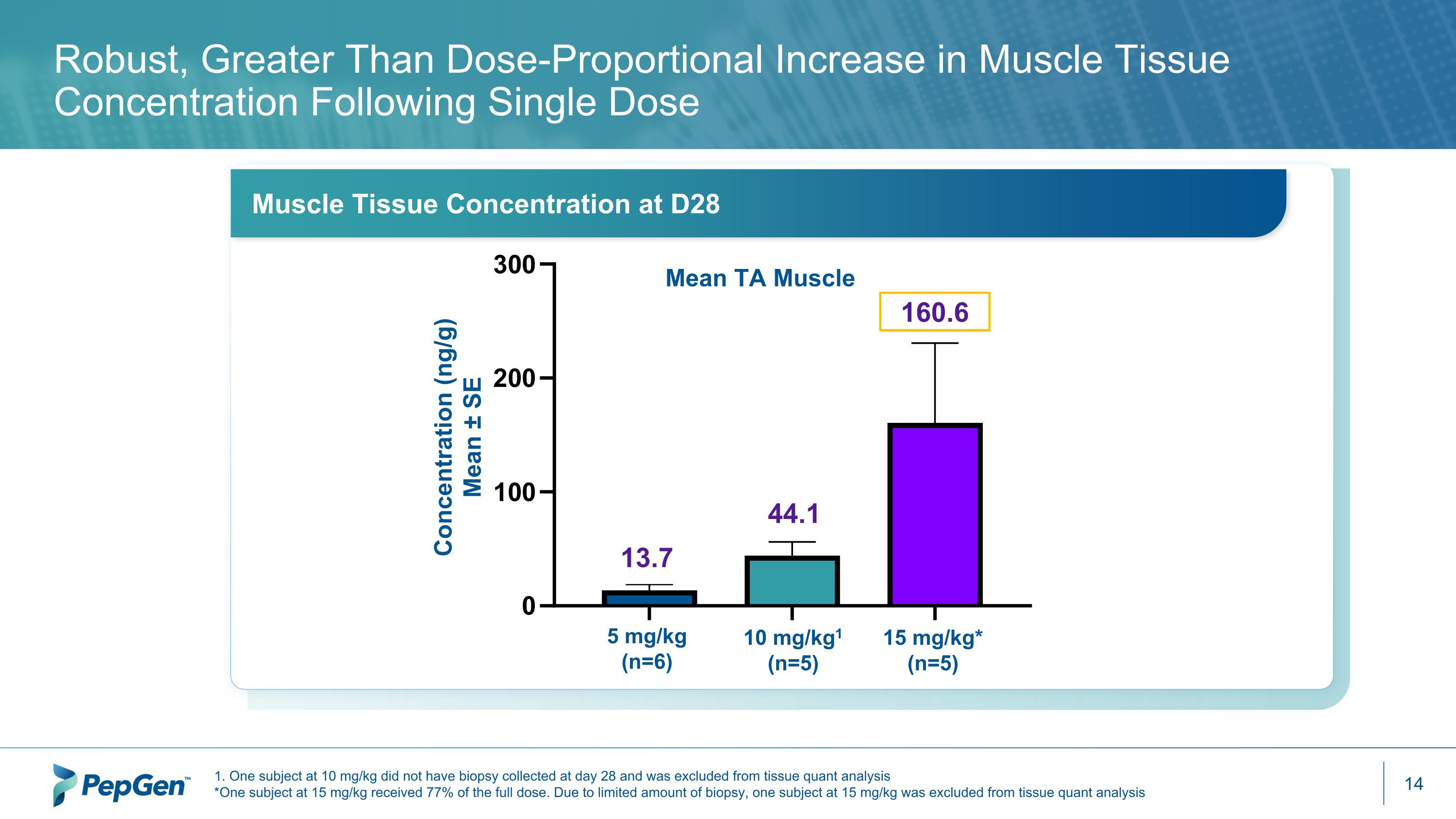
1. One subject at 10 mg/kg did not have biopsy collected at day 28 and was excluded from tissue quant analysis *One subject at 15 mg/kg received 77% of the full dose. Due to limited amount of biopsy, one subject at 15 mg/kg was excluded from tissue quant analysis * Robust, Greater Than Dose-Proportional Increase in Muscle Tissue Concentration Following Single Dose 5 mg/kg (n=6) 10 mg/kg1 (n=5) 15 mg/kg* (n=5) Concentration (ng/g) Mean ± SE 160.6 44.1 13.7 Muscle Tissue Concentration at D28 Mean TA Muscle
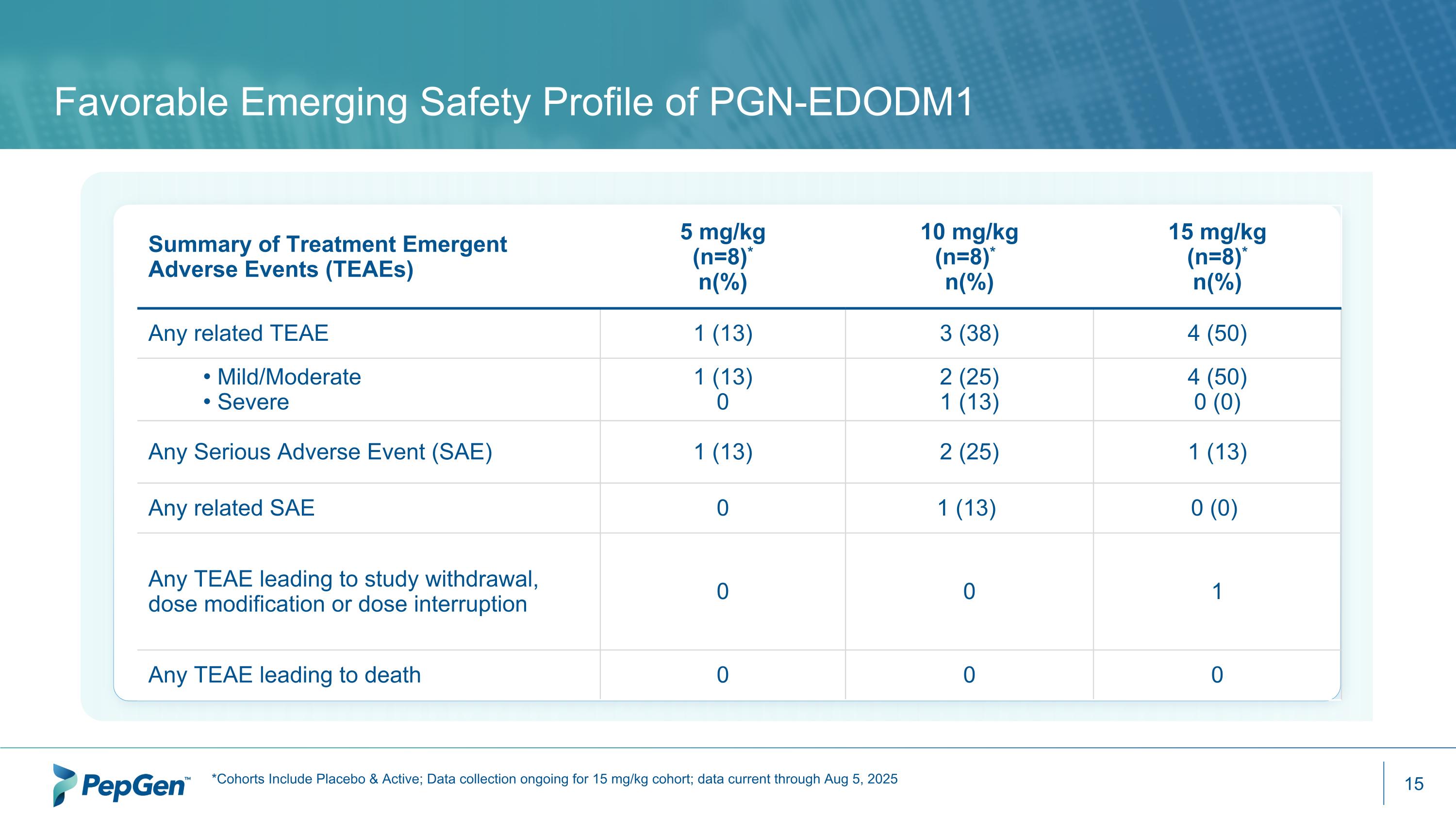
Favorable Emerging Safety Profile of PGN-EDODM1 Summary of Treatment Emergent Adverse Events (TEAEs) 5 mg/kg (n=8)* n(%) 10 mg/kg (n=8)*1 n(%) 15 mg/kg (n=8)* n(%) Any related TEAE 1 (13) 3 (38) 4 (50) Mild/Moderate Severe 1 (13) 0 2 (25) 1 (13) 4 (50) 0 (0) Any Serious Adverse Event (SAE) 1 (13) 2 (25) 1 (13) Any related SAE 0 1 (13) 0 (0) Any TEAE leading to study withdrawal, dose modification or dose interruption 0 0 1 Any TEAE leading to death 0 0 0 *Cohorts Include Placebo & Active; Data collection ongoing for 15 mg/kg cohort; data current through Aug 5, 2025
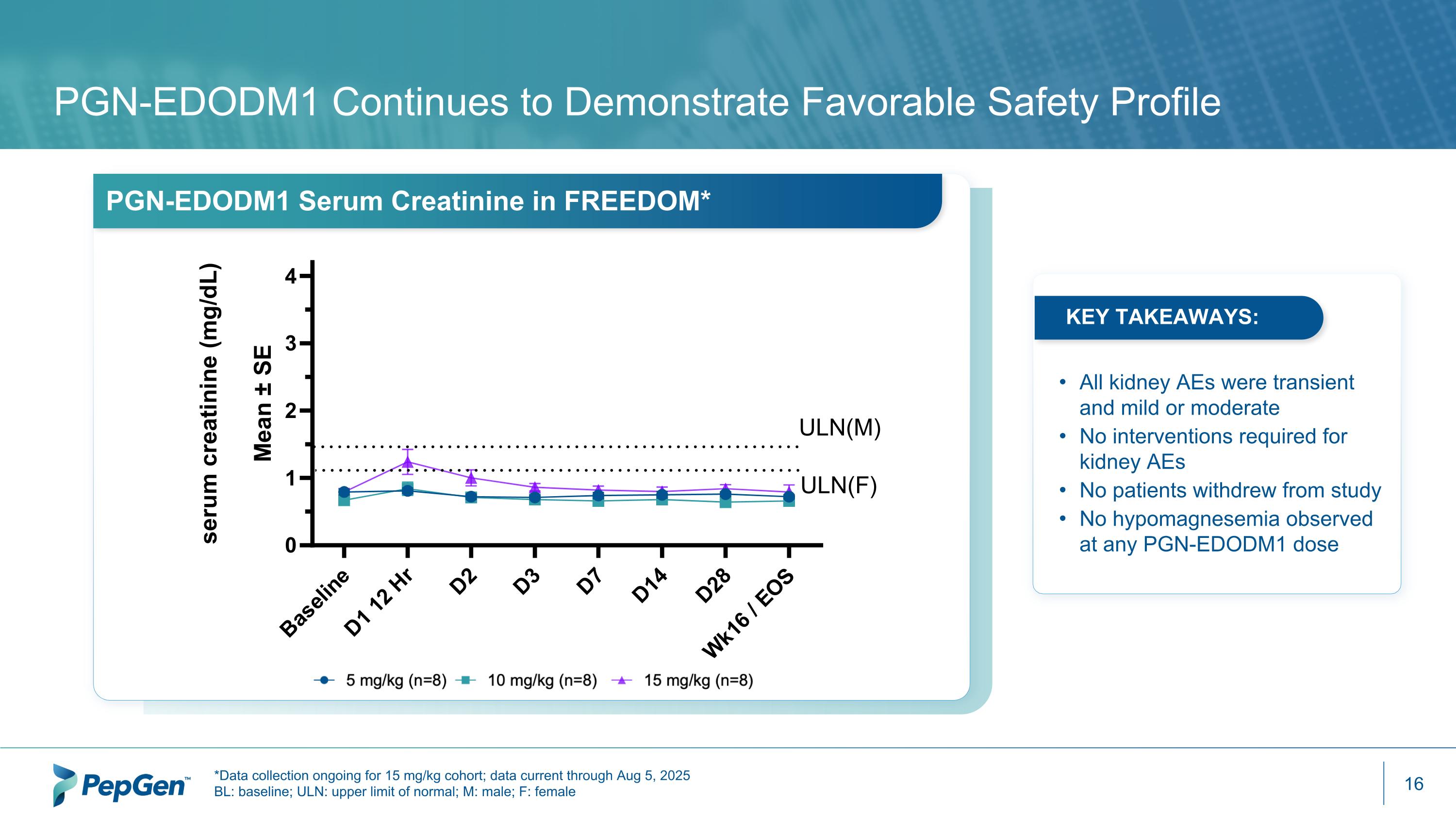
PGN-EDODM1 Continues to Demonstrate Favorable Safety Profile PGN-EDODM1 Serum Creatinine in FREEDOM* *Data collection ongoing for 15 mg/kg cohort; data current through Aug 5, 2025 BL: baseline; ULN: upper limit of normal; M: male; F: female All kidney AEs were transient and mild or moderate No interventions required for kidney AEs No patients withdrew from study No hypomagnesemia observed at any PGN-EDODM1 dose KEY TAKEAWAYS:
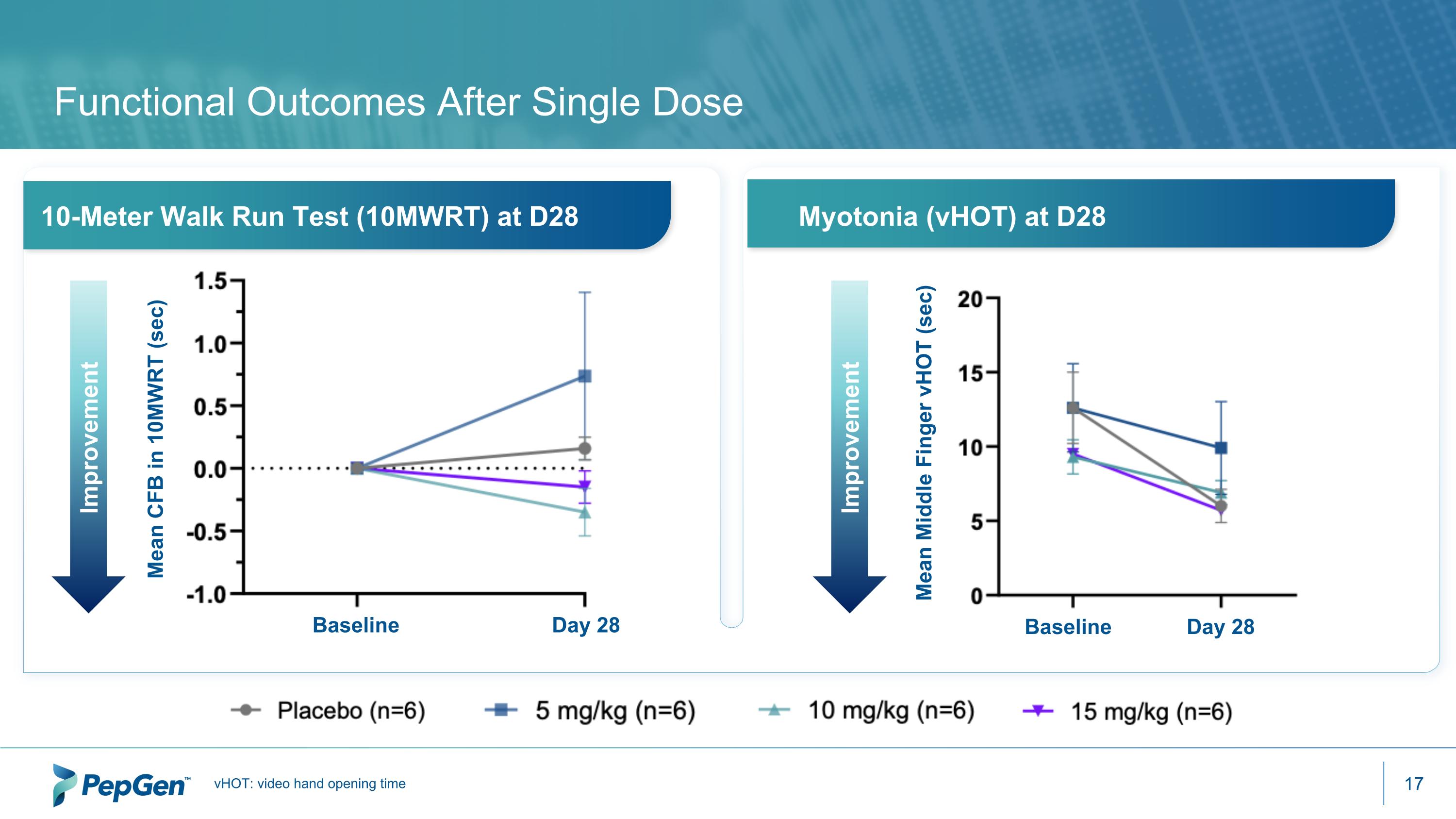
Functional Outcomes After Single Dose vHOT: video hand opening time 10-Meter Walk Run Test (10MWRT) at D28 Myotonia (vHOT) at D28 Baseline Day 28 Mean CFB in 10MWRT (sec) Improvement Improvement Baseline Day 28 Mean Middle Finger vHOT (sec)
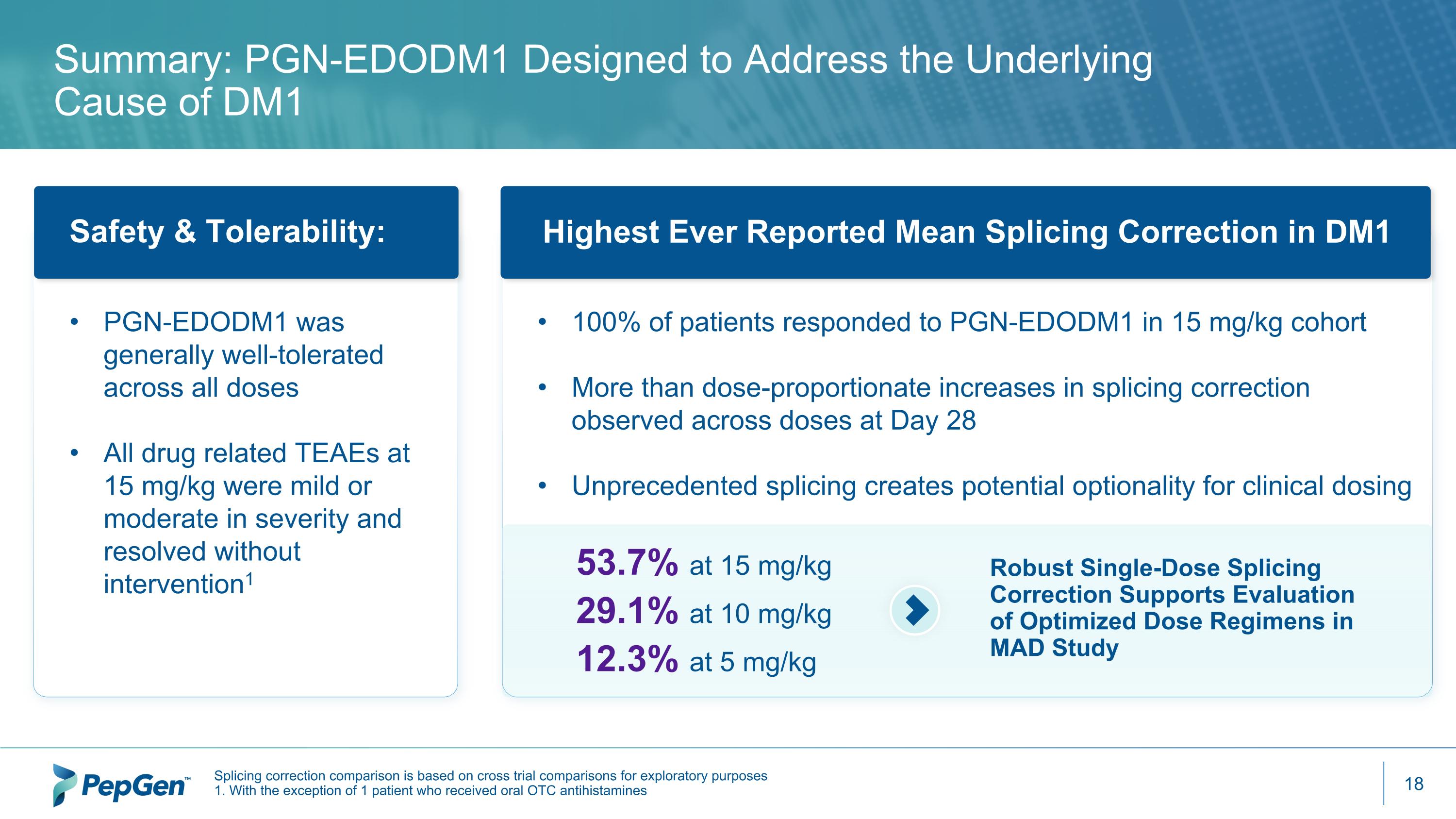
Summary: PGN-EDODM1 Designed to Address the Underlying Cause of DM1 100% of patients responded to PGN-EDODM1 in 15 mg/kg cohort More than dose-proportionate increases in splicing correction observed across doses at Day 28 Unprecedented splicing creates potential optionality for clinical dosing Highest Ever Reported Mean Splicing Correction in DM1 53.7% at 15 mg/kg 29.1% at 10 mg/kg 12.3% at 5 mg/kg Robust Single-Dose Splicing Correction Supports Evaluation of Optimized Dose Regimens in MAD Study Safety & Tolerability: PGN-EDODM1 was generally well-tolerated across all doses All drug related TEAEs at 15 mg/kg were mild or moderate in severity and resolved without intervention1 Splicing correction comparison is based on cross trial comparisons for exploratory purposes 1. With the exception of 1 patient who received oral OTC antihistamines
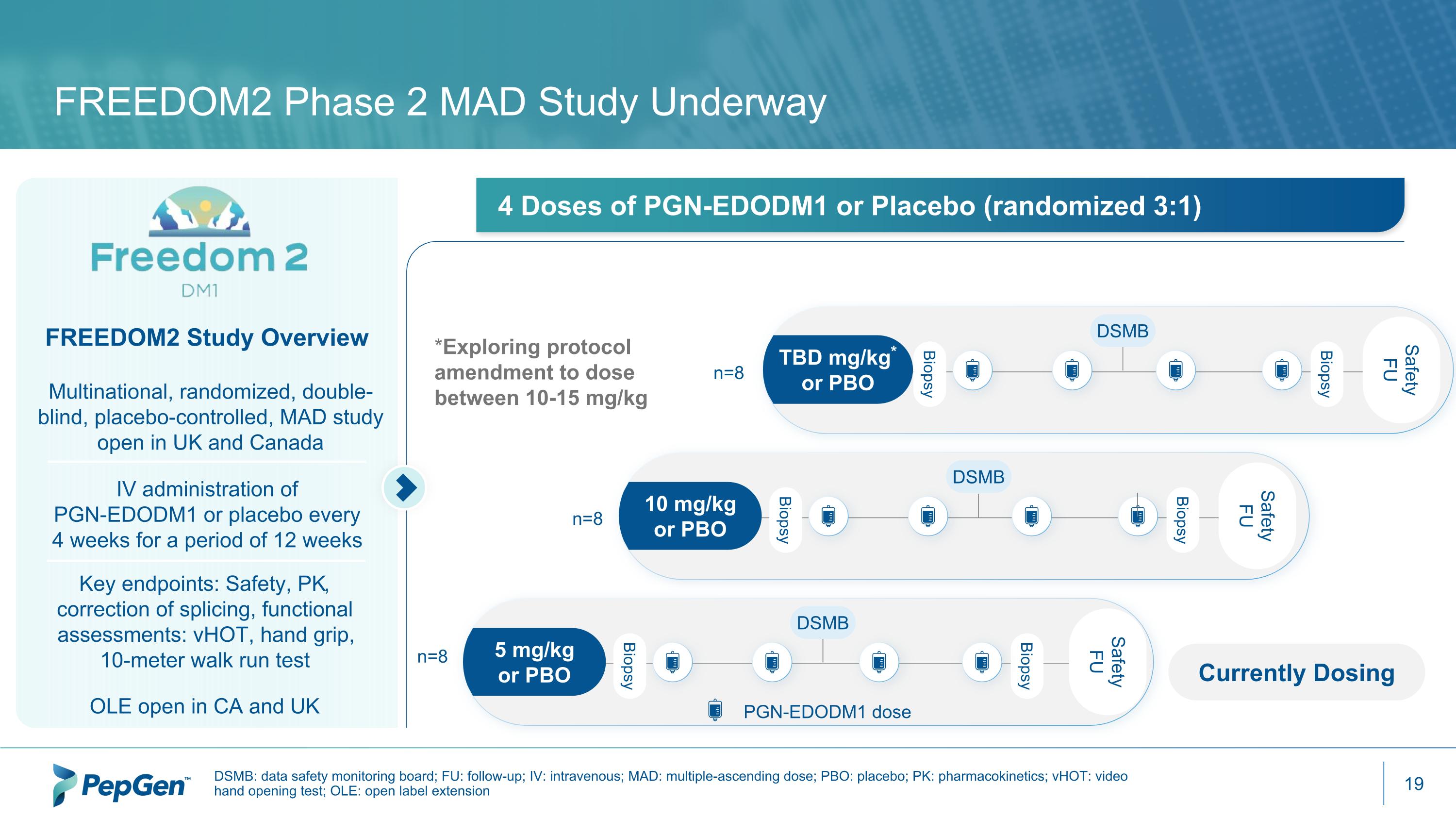
FREEDOM2 Phase 2 MAD Study Underway DSMB: data safety monitoring board; FU: follow-up; IV: intravenous; MAD: multiple-ascending dose; PBO: placebo; PK: pharmacokinetics; vHOT: video hand opening test; OLE: open label extension FREEDOM2 Study Overview Multinational, randomized, double-blind, placebo-controlled, MAD study open in UK and Canada IV administration of PGN-EDODM1 or placebo every 4 weeks for a period of 12 weeks OLE open in CA and UK 4 Doses of PGN-EDODM1 or Placebo (randomized 3:1) n=8 *Exploring protocol amendment to dose between 10-15 mg/kg Safety FU Biopsy Biopsy TBD mg/kg* or PBO DSMB Safety FU Biopsy Biopsy 10 mg/kg or PBO DSMB Biopsy Biopsy 5 mg/kg or PBO DSMB n=8 n=8 Safety FU Currently Dosing PGN-EDODM1 dose Key endpoints: Safety, PK, correction of splicing, functional assessments: vHOT, hand grip, 10-meter walk run test
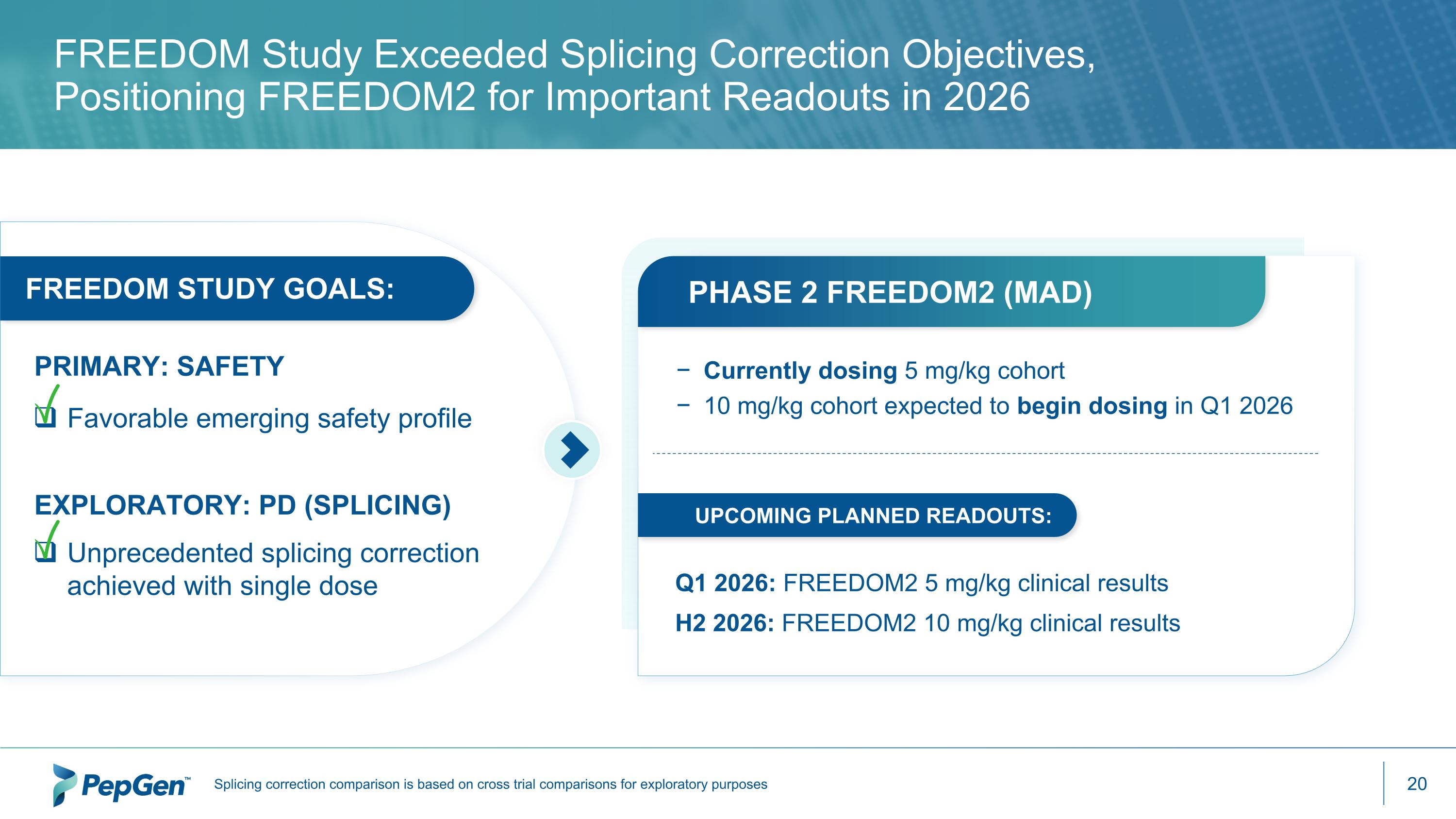
FREEDOM Study Exceeded Splicing Correction Objectives, Positioning FREEDOM2 for Important Readouts in 2026 Favorable emerging safety profile Unprecedented splicing correction achieved with single dose FREEDOM STUDY GOALS: EXPLORATORY: PD (SPLICING) PRIMARY: SAFETY Currently dosing 5 mg/kg cohort 10 mg/kg cohort expected to begin dosing in Q1 2026 PHASE 2 FREEDOM2 (MAD) UPCOMING PLANNED READOUTS: Q1 2026: FREEDOM2 5 mg/kg clinical results H2 2026: FREEDOM2 10 mg/kg clinical results Splicing correction comparison is based on cross trial comparisons for exploratory purposes
