

.3 Delivering on the promise of Prime Editing Corporate Presentation May 2025 1

Forward Looking Statements This presentation contains forward-looking statements of Prime Medicine, Inc. ( Prime , we or our ) within the meaning of the Private Securities Litigation Reform Act of 1995, as amended. These forward-looking statements contain information about our current and future prospects and our operations, which are based on currently available information. All statements other than statements of historical facts contained in this presentation, including statements regarding our strategy, projects and plans are forward-looking statements. In some cases, you can identify forward-looking statements by terminology such as “aim,” “anticipate,” “assume,” “believe,” “contemplate,” “continue” “could,” “design,” “due,” “estimate,” “expect,” “goal,” “hope,” “intend,” “may,” “might,” “objective,” “opportunity,” “plan,” “predict,” “positioned,” “possible,” “potential,” “project,” “seek,” “should,” “strategy,” “target,” “will,” “would” and other similar expressions that are predictions of or indicate future events and future trends, or the negative of these terms or other comparable terminology. These forward-looking statements include, but are not limited to, express or implied statements about Prime’s beliefs and expectations regarding: the potential of Prime Editing to correct the causative mutations of diseases, including CGD, Wilson’s Disease, CF, and AATD; the continued development and advancement of its AATD and Wilson’s Disease programs, including the timing of the filing of IND and/or CTA applications in mid-2026 and 1H 2026, respectively, and the timing of initial data for both programs in 2027; the initiation, timing, progress and results of our research and development programs, preclinical studies and clinical trials, including the release of data; the safety profile of Prime Editing, our modular LNP, and our programs; our ability to launch therapeutics; the timing of, and our ability to achieve, clinical validation and sustained, long-term value creation; the modularity of the Prime Editing platform and the benefits thereof; the collaboration with Bristol Myers Squibb and the intended and potential benefits thereof, including the receipt of potential milestone and royalty payments from commercial product sales, if any; our expectations regarding the breadth of Prime Editing, including the potential of Prime Editing to address more than 90% of genetic diseases and to address non-genetic diseases; the continued development and optimization of various non-viral and viral delivery systems, including our universal liver-targeted LNP delivery approach; the scope of protection we are able to establish and maintain for intellectual property rights covering our Prime Editing technology; the implementation of our strategic plans for our business, programs and technology, including our ability to identify and enter into future license agreements and collaborations; regulatory developments in the United States and foreign countries; our ability to attract and retain key scientific and management personnel; our estimates of our expenses, capital requirements, and needs for additional financing; and our expectations regarding the anticipated timeline of our cash runway and future financial performance. Actual results or events could differ materially from the plans, intentions and expectations disclosed in the forward-looking statements we make due to a number of risks and uncertainties. These and other risks, uncertainties and important factors are described in the section entitled Risk Factors in our most recent Annual Report on Form 10-K, as well as any subsequent filings with the Securities and Exchange Commission. Any forward-looking statements represent our views only as of the date of this presentation and we undertake no obligation to update or revise any forward-looking statements, whether as a result of new information, the occurrence of certain events or otherwise subject to any obligations under applicable law. We may not actually achieve the plans, intentions or expectations disclosed in our forward-looking statements, and you should not place undue reliance on our forward-looking statements. No representations or warranties (expressed or implied) are made about the accuracy of any such forward-looking statements. Certain information contained in this presentation relates to or is based on studies, publications, surveys and other data obtained from third-party sources and our own internal estimates and research. While we believe these third-party studies, publications, surveys and other data to be reliable as of the date of this presentation, we have not independently verified, and make no representation as to the adequacy, fairness, accuracy or completeness of, any information obtained from third-party sources. In addition, no independent source has evaluated the reasonableness or accuracy of our internal estimates or research and no reliance should be made on any information or statements made in this presentation relating to or based on such internal estimates and research. 2

We are advancing Prime Editing to change the course of how diseases are treated. We aim to provide safe, effective and curative treatments, which offer lifelong benefit to patients. 3
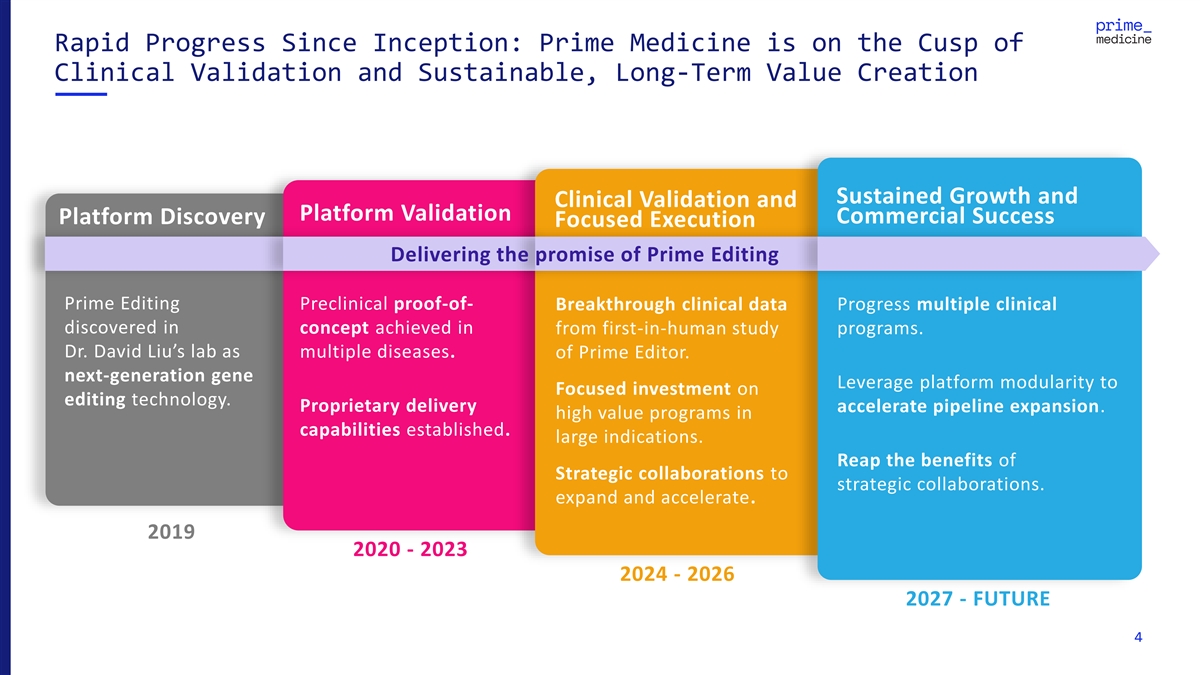
Rapid Progress Since Inception: Prime Medicine is on the Cusp of Clinical Validation and Sustainable, Long-Term Value Creation Sustained Growth and Clinical Validation and Platform Validation Commercial Success Platform Discovery Focused Execution Delivering the promise of Prime Editing Prime Editing Preclinical proof-of- Breakthrough clinical data Progress multiple clinical discovered in concept achieved in from first-in-human study programs. Dr. David Liu’s lab as multiple diseases. of Prime Editor. next-generation gene Leverage platform modularity to Focused investment on editing technology. Proprietary delivery accelerate pipeline expansion. high value programs in capabilities established. large indications. Reap the benefits of Strategic collaborations to strategic collaborations. expand and accelerate. 2019 2020 - 2023 2024 - 2026 2027 - FUTURE 4
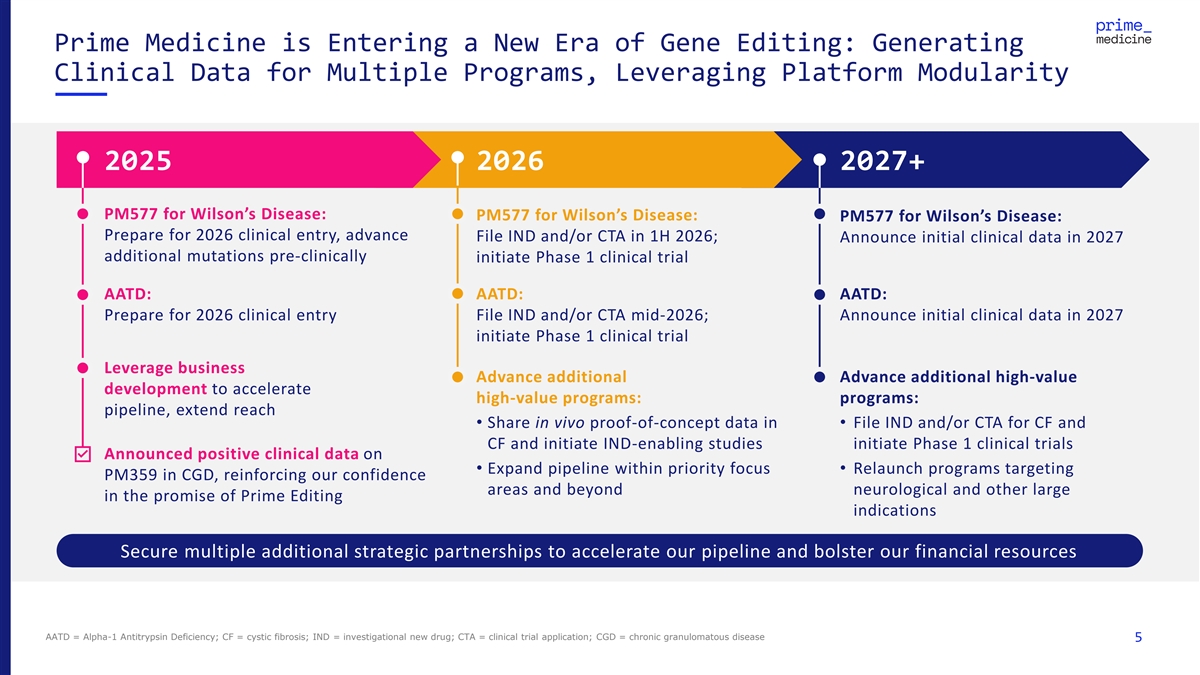
Prime Medicine is Entering a New Era of Gene Editing: Generating Clinical Data for Multiple Programs, Leveraging Platform Modularity 2025 2026 2027+ PM577 for Wilson’s Disease: PM577 for Wilson’s Disease: PM577 for Wilson’s Disease: Prepare for 2026 clinical entry, advance File IND and/or CTA in 1H 2026; Announce initial clinical data in 2027 additional mutations pre-clinically initiate Phase 1 clinical trial AATD: AATD: AATD: Prepare for 2026 clinical entry File IND and/or CTA mid-2026; Announce initial clinical data in 2027 initiate Phase 1 clinical trial Leverage business Advance additional Advance additional high-value development to accelerate high-value programs: programs: pipeline, extend reach • Share in vivo proof-of-concept data in • File IND and/or CTA for CF and CF and initiate IND-enabling studies initiate Phase 1 clinical trials Announced positive clinical data on • Expand pipeline within priority focus • Relaunch programs targeting PM359 in CGD, reinforcing our confidence areas and beyond neurological and other large in the promise of Prime Editing indications Secure multiple additional strategic partnerships to accelerate our pipeline and bolster our financial resources AATD = Alpha-1 Antitrypsin Deficiency; CF = cystic fibrosis; IND = investigational new drug; CTA = clinical trial application; CGD = chronic granulomatous disease 5
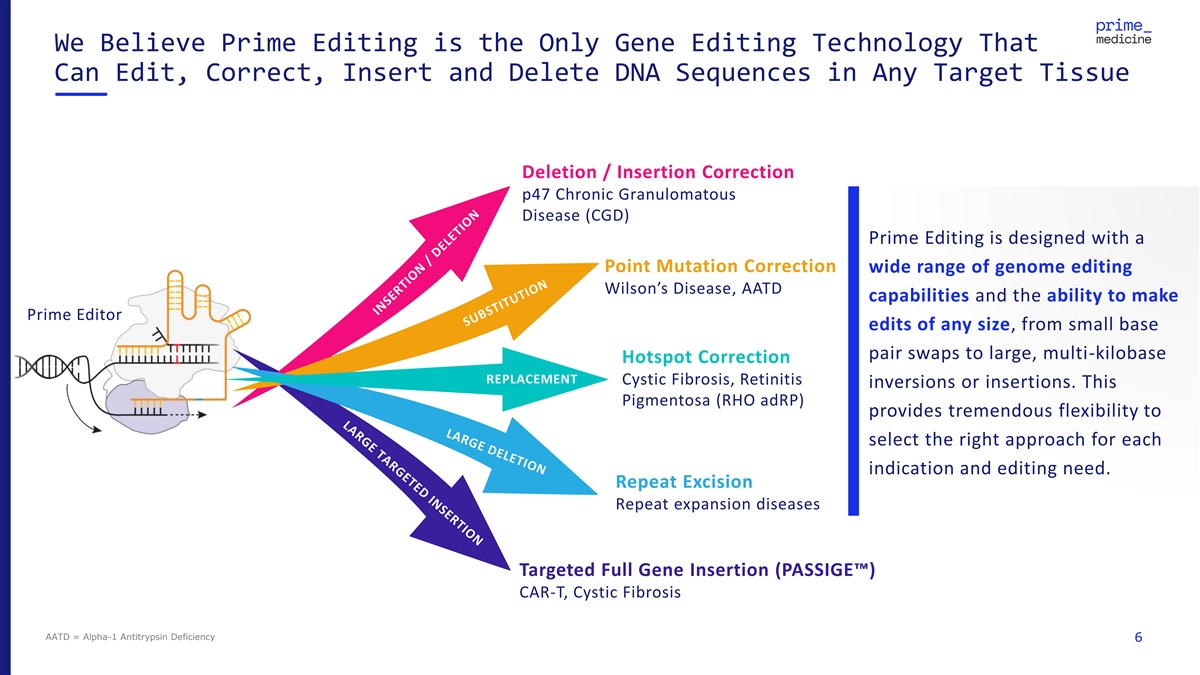
We Believe Prime Editing is the Only Gene Editing Technology That Can Edit, Correct, Insert and Delete DNA Sequences in Any Target Tissue Deletion / Insertion Correction p47 Chronic Granulomatous Disease (CGD) Prime Editing is designed with a Point Mutation Correction wide range of genome editing Wilson’s Disease, AATD capabilities and the ability to make Prime Editor edits of any size, from small base pair swaps to large, multi-kilobase Hotspot Correction REPLACEMENT Cystic Fibrosis, Retinitis inversions or insertions. This Pigmentosa (RHO adRP) provides tremendous flexibility to select the right approach for each indication and editing need. Repeat Excision Repeat expansion diseases Targeted Full Gene Insertion (PASSIGE ) CAR-T, Cystic Fibrosis AATD = Alpha-1 Antitrypsin Deficiency 6
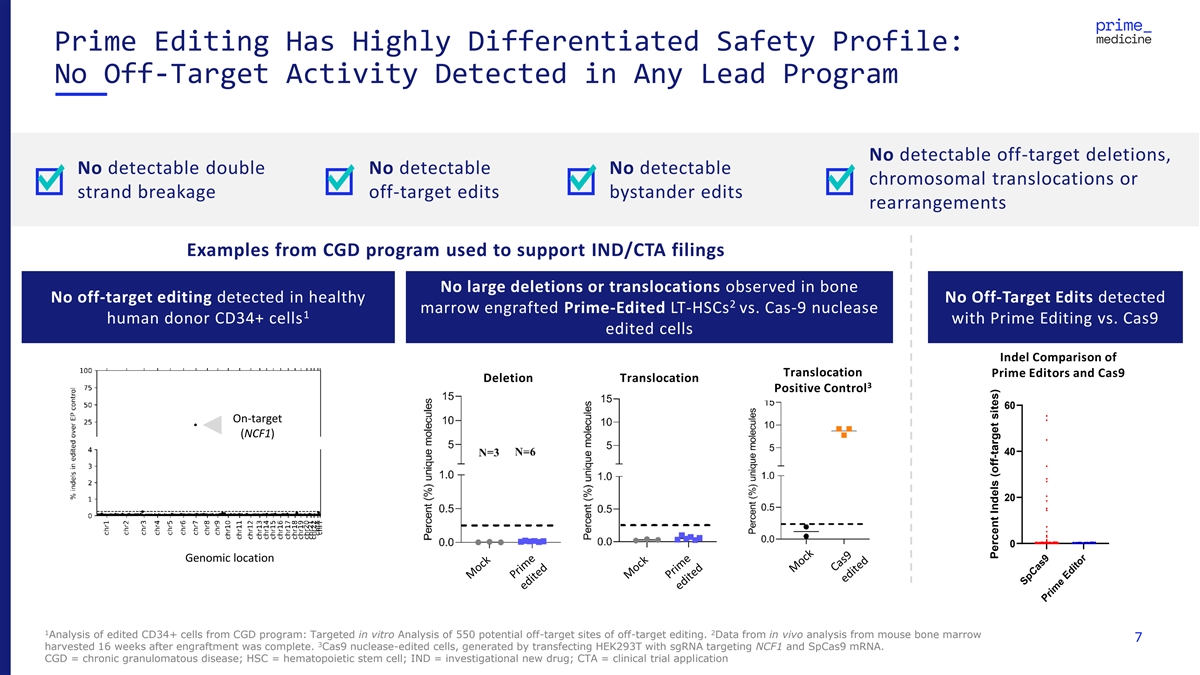
Prime Editing Has Highly Differentiated Safety Profile: No Off-Target Activity Detected in Any Lead Program No detectable off-target deletions, No detectable double No detectable No detectable chromosomal translocations or strand breakage off-target edits bystander edits rearrangements Examples from CGD program used to support IND/CTA filings No large deletions or translocations observed in bone No off-target editing detected in healthy No Off-Target Edits detected 2 marrow engrafted Prime-Edited LT-HSCs vs. Cas-9 nuclease 1 human donor CD34+ cells with Prime Editing vs. Cas9 edited cells Indel Comparison of Translocation Prim 12 e E Spa dito cer rs a snd Cas9 Deletion Translocation 3 Positive Control 60 On-target (NCF1) 40 20 0 Genomic location 1 2 Analysis of edited CD34+ cells from CGD program: Targeted in vitro Analysis of 550 potential off-target sites of off-target editing. Data from in vivo analysis from mouse bone marrow 7 3 harvested 16 weeks after engraftment was complete. Cas9 nuclease-edited cells, generated by transfecting HEK293T with sgRNA targeting NCF1 and SpCas9 mRNA. CGD = chronic granulomatous disease; HSC = hematopoietic stem cell; IND = investigational new drug; CTA = clinical trial application SpCas9 Prime Editor Percent Indels (off-target sites)
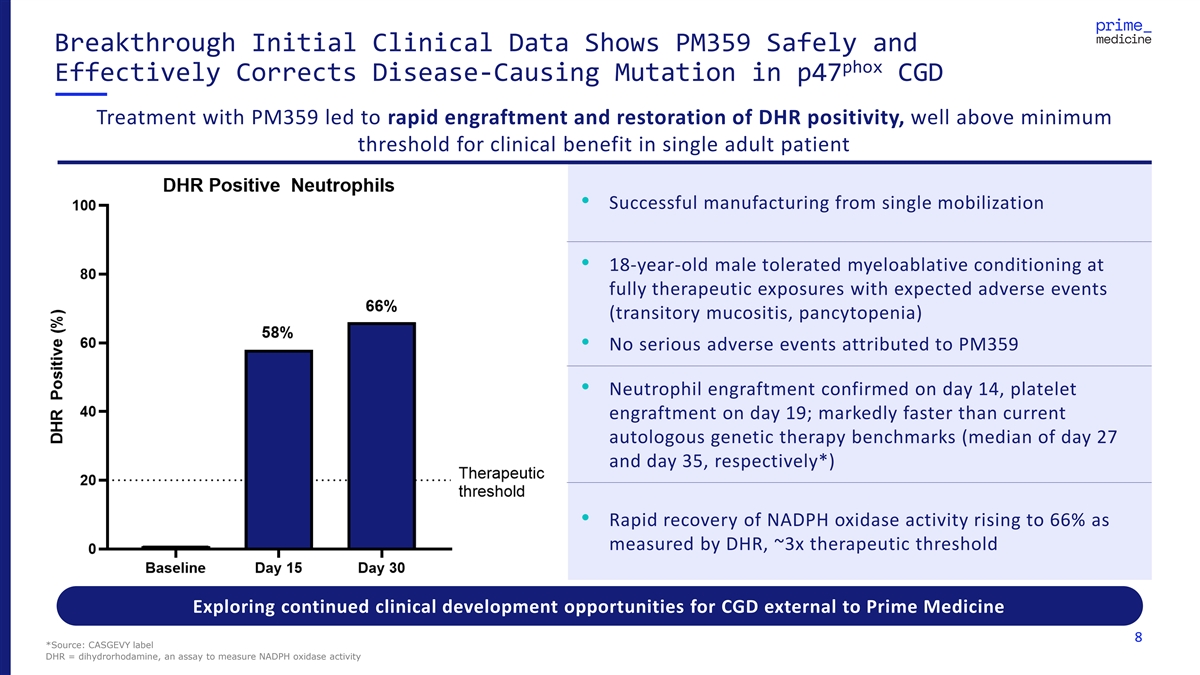
Breakthrough Initial Clinical Data Shows PM359 Safely and phox Effectively Corrects Disease-Causing Mutation in p47 CGD Treatment with PM359 led to rapid engraftment and restoration of DHR positivity, well above minimum threshold for clinical benefit in single adult patient • Successful manufacturing from single mobilization • 18-year-old male tolerated myeloablative conditioning at fully therapeutic exposures with expected adverse events (transitory mucositis, pancytopenia) • No serious adverse events attributed to PM359 • Neutrophil engraftment confirmed on day 14, platelet engraftment on day 19; markedly faster than current autologous genetic therapy benchmarks (median of day 27 and day 35, respectively*) • Rapid recovery of NADPH oxidase activity rising to 66% as measured by DHR, ~3x therapeutic threshold Exploring continued clinical development opportunities for CGD external to Prime Medicine 8 *Source: CASGEVY label DHR = dihydrorhodamine, an assay to measure NADPH oxidase activity
Prime Editing Platform Modularity Accelerates and De-Risks Ongoing Efforts, Enables Rapid Generation of New Product Candidates Off-Target Assays Prime Editors Manufacturing Clinical Delivery Nonclinical Regulatory 9
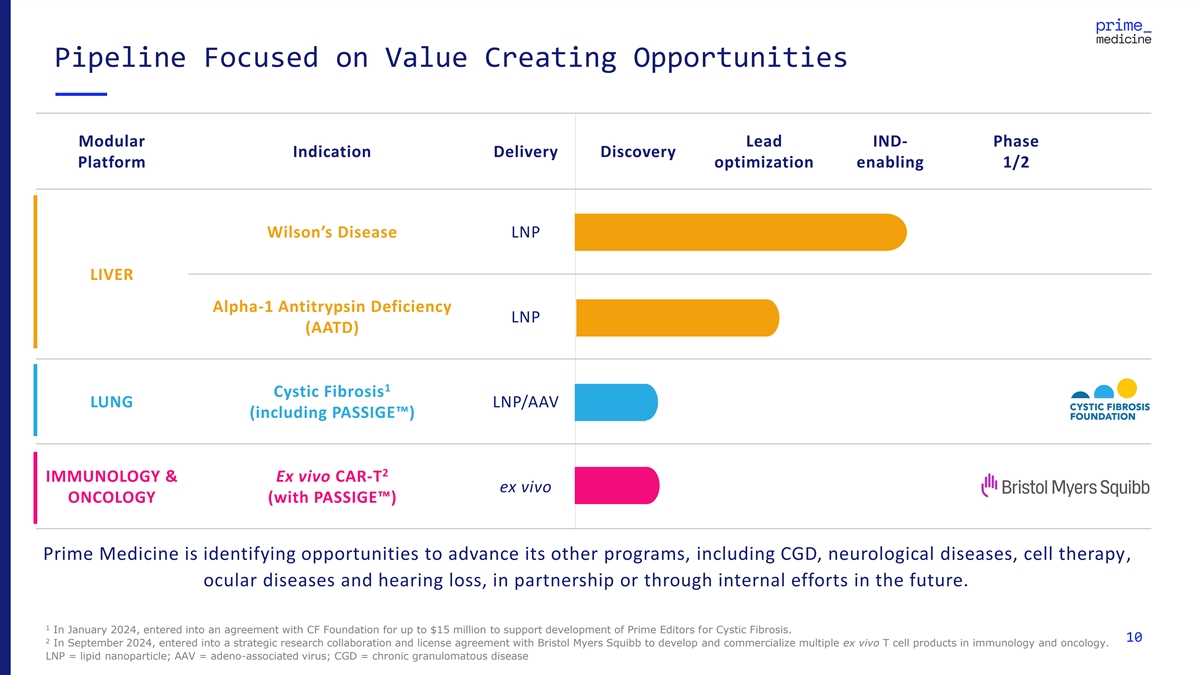
Pipeline Focused on Value Creating Opportunities Modular Lead IND- Phase Indication Delivery Discovery Platform optimization enabling 1/2 Wilson’s Disease LNP LIVER Alpha-1 Antitrypsin Deficiency LNP (AATD) 1 Cystic Fibrosis LUNG LNP/AAV (including PASSIGE ) 2 IMMUNOLOGY & Ex vivo CAR-T ex vivo ONCOLOGY (with PASSIGE ) Prime Medicine is identifying opportunities to advance its other programs, including CGD, neurological diseases, cell therapy, ocular diseases and hearing loss, in partnership or through internal efforts in the future. 1 In January 2024, entered into an agreement with CF Foundation for up to $15 million to support development of Prime Editors for Cystic Fibrosis. 10 2 In September 2024, entered into a strategic research collaboration and license agreement with Bristol Myers Squibb to develop and commercialize multiple ex vivo T cell products in immunology and oncology. LNP = lipid nanoparticle; AAV = adeno-associated virus; CGD = chronic granulomatous disease

Liver | 11 11
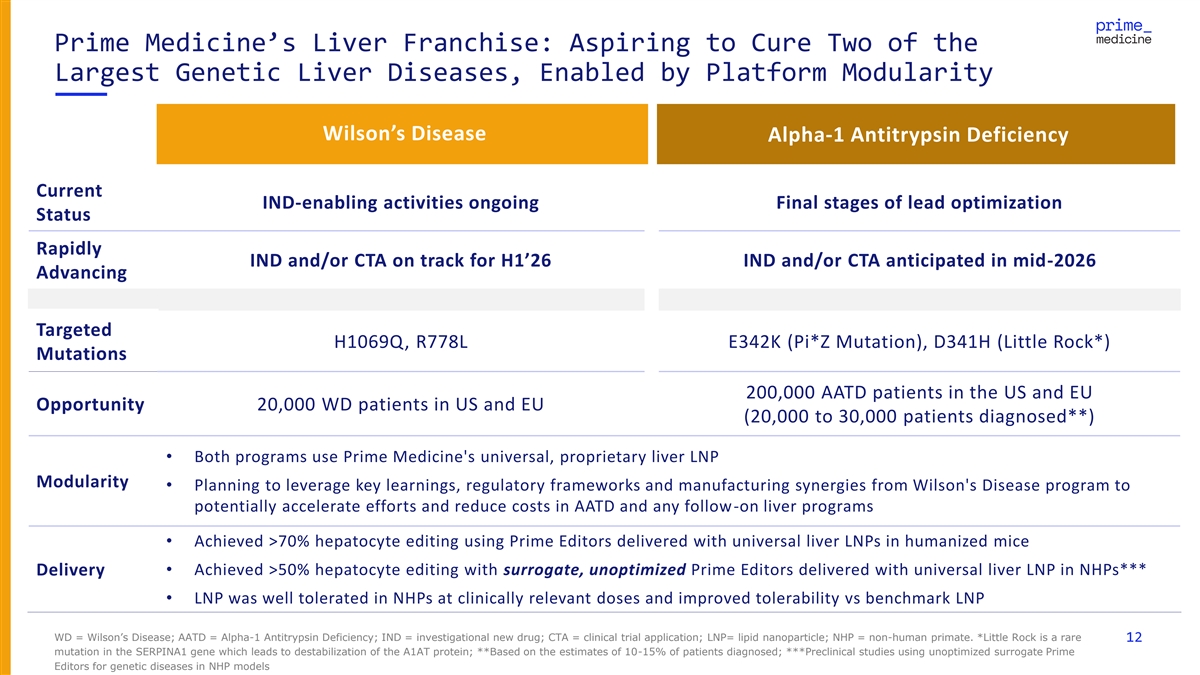
Prime Medicine’s Liver Franchise: Aspiring to Cure Two of the Largest Genetic Liver Diseases, Enabled by Platform Modularity Wilson’s Disease Alpha-1 Antitrypsin Deficiency Current IND-enabling activities ongoing Final stages of lead optimization Status Rapidly IND and/or CTA on track for H1’26 IND and/or CTA anticipated in mid-2026 Advancing Targeted H1069Q, R778L E342K (Pi*Z Mutation), D341H (Little Rock*) Mutations 200,000 AATD patients in the US and EU Opportunity 20,000 WD patients in US and EU (20,000 to 30,000 patients diagnosed**) • Both programs use Prime Medicine's universal, proprietary liver LNP Modularity • Planning to leverage key learnings, regulatory frameworks and manufacturing synergies from Wilson's Disease program to potentially accelerate efforts and reduce costs in AATD and any follow-on liver programs • Achieved >70% hepatocyte editing using Prime Editors delivered with universal liver LNPs in humanized mice • Achieved >50% hepatocyte editing with surrogate, unoptimized Prime Editors delivered with universal liver LNP in NHPs*** Delivery • LNP was well tolerated in NHPs at clinically relevant doses and improved tolerability vs benchmark LNP WD = Wilson’s Disease; AATD = Alpha-1 Antitrypsin Deficiency; IND = investigational new drug; CTA = clinical trial application; LNP= lipid nanoparticle; NHP = non-human primate. *Little Rock is a rare 12 mutation in the SERPINA1 gene which leads to destabilization of the A1AT protein; **Based on the estimates of 10-15% of patients diagnosed; ***Preclinical studies using unoptimized surrogate Prime Editors for genetic diseases in NHP models
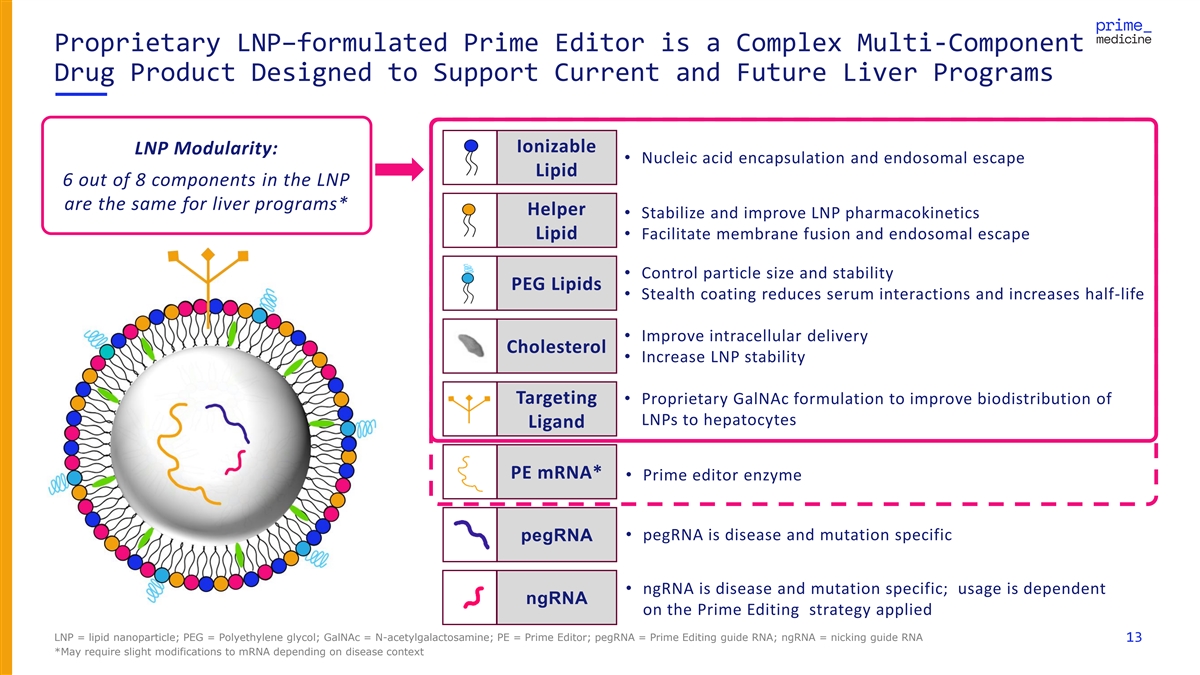
Proprietary LNP–formulated Prime Editor is a Complex Multi-Component Drug Product Designed to Support Current and Future Liver Programs Ionizable LNP Modularity: • Nucleic acid encapsulation and endosomal escape Lipid 6 out of 8 components in the LNP are the same for liver programs* Helper • Stabilize and improve LNP pharmacokinetics Lipid• Facilitate membrane fusion and endosomal escape • Control particle size and stability PEG Lipids • Stealth coating reduces serum interactions and increases half-life • Improve intracellular delivery Cholesterol • Increase LNP stability Targeting • Proprietary GalNAc formulation to improve biodistribution of LNPs to hepatocytes Ligand PE mRNA* • Prime editor enzyme • pegRNA is disease and mutation specific pegRNA • ngRNA is disease and mutation specific; usage is dependent ngRNA on the Prime Editing strategy applied LNP = lipid nanoparticle; PEG = Polyethylene glycol; GalNAc = N-acetylgalactosamine; PE = Prime Editor; pegRNA = Prime Editing guide RNA; ngRNA = nicking guide RNA 13 *May require slight modifications to mRNA depending on disease context
Prime Medicine’s Modular LNP Dosed as a Single Administration in Cynomolgus Monkey (NHP) With Minimal, If Any Safety Concerns • Well-tolerated: no acute reactions, clinical observations, or body weight changes • Transient LFT elevations • No change in platelets, coagulation or blood count • No change in blood biochemistry • Minimal changes in IL6 levels • No other cytokine changes • No change in liver histopathology (H&E) • Animals healthy at 44 weeks • Benchmarked favorably against other LNPs in clinical development LNP = lipid nanoparticle; LFT = Liver Function Tests; H&E = hematoxylin and eosin 14

Wilson’s Disease Confidential | 15 15
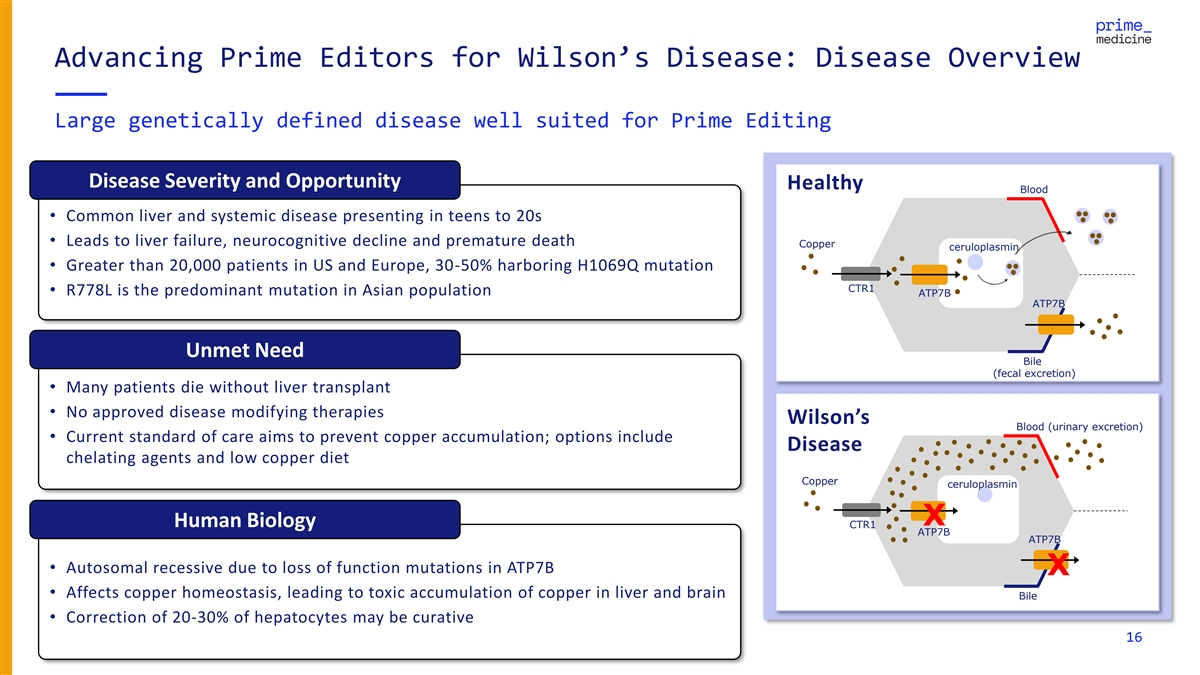
Advancing Prime Editors for Wilson’s Disease: Disease Overview Large genetically defined disease well suited for Prime Editing Disease Severity and Opportunity Healthy Blood • Common liver and systemic disease presenting in teens to 20s • Leads to liver failure, neurocognitive decline and premature death Copper ceruloplasmin • Greater than 20,000 patients in US and Europe, 30-50% harboring H1069Q mutation CTR1 • R778L is the predominant mutation in Asian population ATP7B ATP7B Unmet Need Bile (fecal excretion) • Many patients die without liver transplant • No approved disease modifying therapies Wilson’s Blood (urinary excretion) • Current standard of care aims to prevent copper accumulation; options include Disease chelating agents and low copper diet Copper ceruloplasmin X Human Biology CTR1 ATP7B ATP7B • Autosomal recessive due to loss of function mutations in ATP7B X • Affects copper homeostasis, leading to toxic accumulation of copper in liver and brain Bile • Correction of 20-30% of hepatocytes may be curative 16
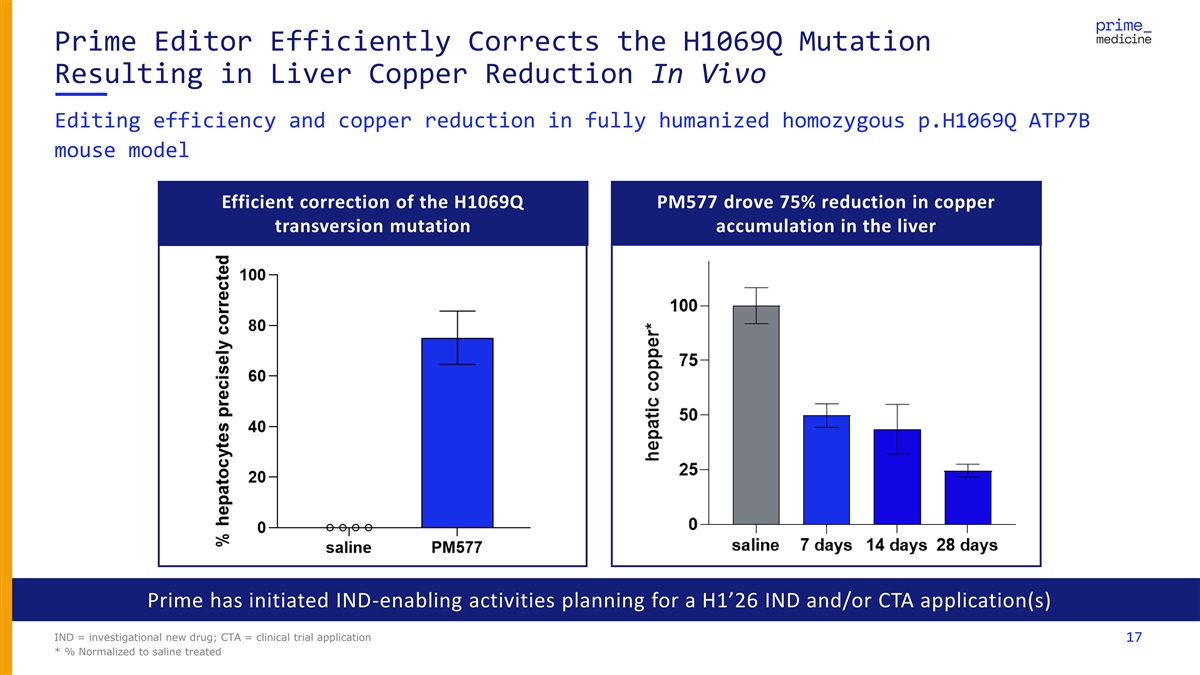
Prime Editor Efficiently Corrects the H1069Q Mutation Resulting in Liver Copper Reduction In Vivo Editing efficiency and copper reduction in fully humanized homozygous p.H1069Q ATP7B mouse model Efficient correction of the H1069Q PM577 drove 75% reduction in copper transversion mutation accumulation in the liver Prime has initiated IND-enabling activities planning for a H1’26 IND and/or CTA application(s) IND = investigational new drug; CTA = clinical trial application 17 * % Normalized to saline treated

Alpha-1 Antitrypsin Deficiency (AATD) Confidential | 18 18

Advancing Prime Editors for AATD: Disease Overview Disease Severity and Opportunity • AATD is an inherited genetic disorder that causes low levels of A1AT protein • Low levels of A1AT protein increases the risk of lung disease (emphysema) • Patients are also at risk of liver disease (cirrhosis) caused by mutant protein aggregation • Approximately 200,000 patients in the US and EU, ~10-15% of which are diagnosed today Unmet Need • Many patients progress to liver failure or severe lung disease, requiring transplant • Current standard of care includes chronic A1AT augmentation therapy for lung disease; no approved curative therapies • No approved treatments for liver disease Human Biology • Autosomal codominant disorder due to mutations in SERPINA1 gene We believe Prime Editing is uniquely • Lung: lack of functional A1AT leads to unrestricted neutrophil elastase activity, among well-suited to correct mutant A1AT other pathological changes (loss of function) • Liver: defective A1AT protein misfolding and accumulation (gain of function) protein to wild-type without the risk • 20-30% correction in hepatocytes could be curative of bystander edits 19 AATD = Alpha-1 Antitrypsin Deficiency; A1AT = alpha-1 antitrypsin
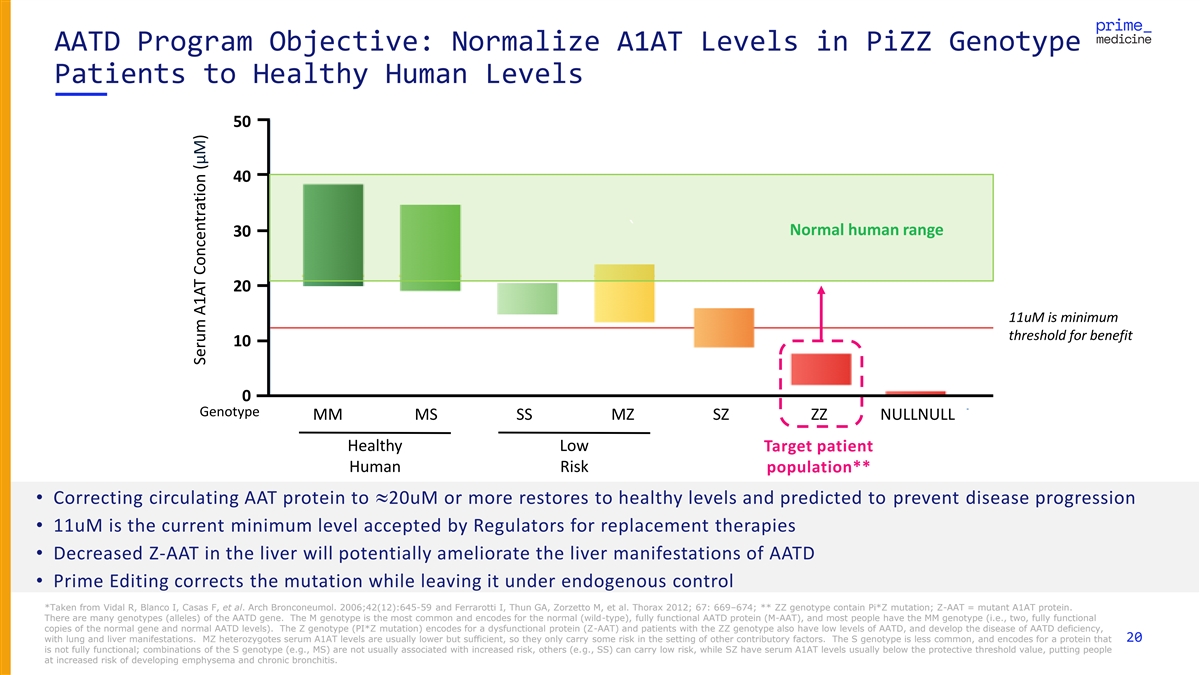
AATD Program Objective: Normalize A1AT Levels in PiZZ Genotype Patients to Healthy Human Levels 50 40 ` Normal human range 30 20 11uM is minimum threshold for benefit 10 0 Genotype MM MS SS MZ SZ ZZ NULLNULL Healthy Low Target patient Human Risk population** • Correcting circulating AAT protein to »20uM or more restores to healthy levels and predicted to prevent disease progression • 11uM is the current minimum level accepted by Regulators for replacement therapies • Decreased Z-AAT in the liver will potentially ameliorate the liver manifestations of AATD • Prime Editing corrects the mutation while leaving it under endogenous control *Taken from Vidal R, Blanco I, Casas F, et al. Arch Bronconeumol. 2006;42(12):645-59 and Ferrarotti I, Thun GA, Zorzetto M, et al. Thorax 2012; 67: 669–674; ** ZZ genotype contain Pi*Z mutation; Z-AAT = mutant A1AT protein. There are many genotypes (alleles) of the AATD gene. The M genotype is the most common and encodes for the normal (wild-type), fully functional AATD protein (M-AAT), and most people have the MM genotype (i.e., two, fully functional copies of the normal gene and normal AATD levels). The Z genotype (PI*Z mutation) encodes for a dysfunctional protein (Z-AAT) and patients with the ZZ genotype also have low levels of AATD, and develop the disease of AATD deficiency, with lung and liver manifestations. MZ heterozygotes serum A1AT levels are usually lower but sufficient, so they only carry some risk in the setting of other contributory factors. The S genotype is less common, and encodes for a protein that 20 is not fully functional; combinations of the S genotype (e.g., MS) are not usually associated with increased risk, others (e.g., SS) can carry low risk, while SZ have serum A1AT levels usually below the protective threshold value, putting people at increased risk of developing emphysema and chronic bronchitis. Serum A1AT Concentration (μM)
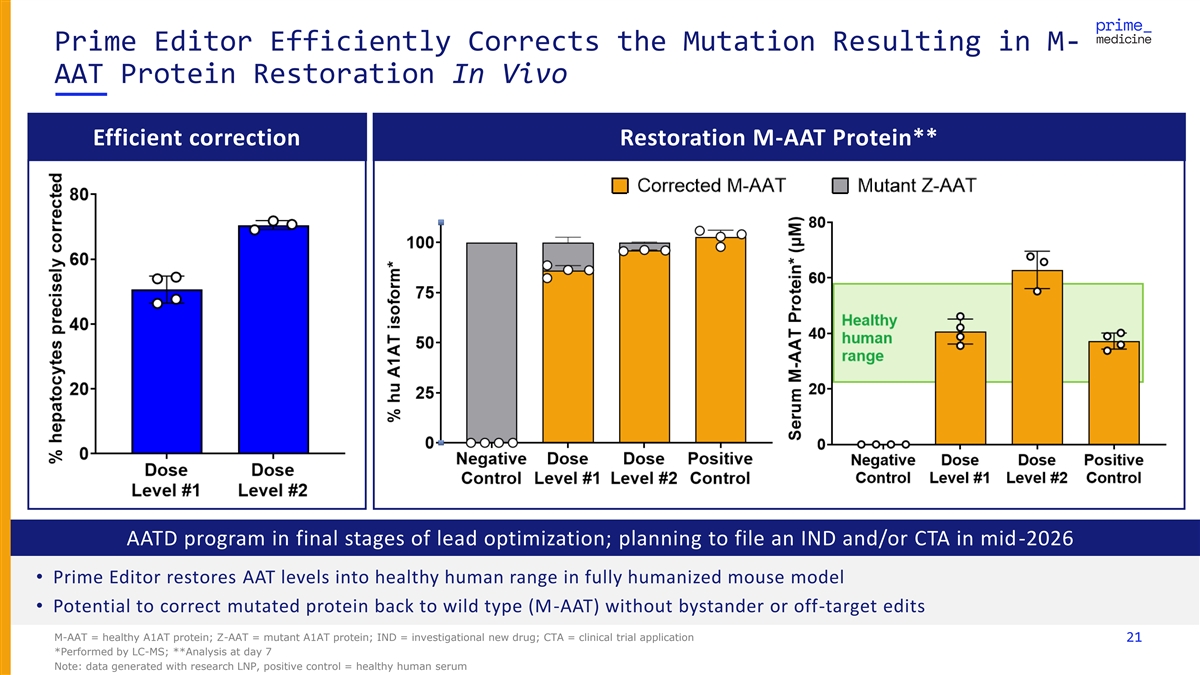
Prime Editor Efficiently Corrects the Mutation Resulting in M- AAT Protein Restoration In Vivo Efficient correction Restoration M-AAT Protein** AATD program in final stages of lead optimization; planning to file an IND and/or CTA in mid-2026 • Prime Editor restores AAT levels into healthy human range in fully humanized mouse model • Potential to correct mutated protein back to wild type (M-AAT) without bystander or off-target edits M-AAT = healthy A1AT protein; Z-AAT = mutant A1AT protein; IND = investigational new drug; CTA = clinical trial application 21 *Performed by LC-MS; **Analysis at day 7 Note: data generated with research LNP, positive control = healthy human serum

Lung | 22 22
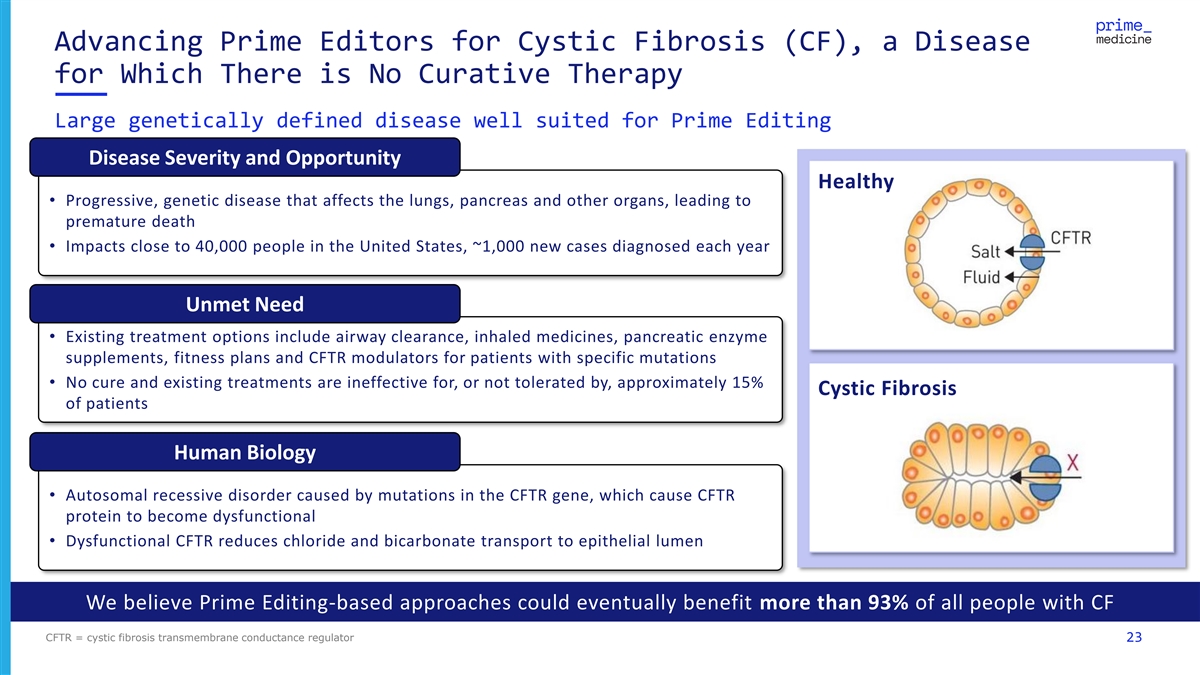
Advancing Prime Editors for Cystic Fibrosis (CF), a Disease for Which There is No Curative Therapy Large genetically defined disease well suited for Prime Editing Disease Severity and Opportunity Healthy • Progressive, genetic disease that affects the lungs, pancreas and other organs, leading to premature death • Impacts close to 40,000 people in the United States, ~1,000 new cases diagnosed each year Unmet Need • Existing treatment options include airway clearance, inhaled medicines, pancreatic enzyme supplements, fitness plans and CFTR modulators for patients with specific mutations • No cure and existing treatments are ineffective for, or not tolerated by, approximately 15% Cystic Fibrosis of patients Human Biology • Autosomal recessive disorder caused by mutations in the CFTR gene, which cause CFTR protein to become dysfunctional • Dysfunctional CFTR reduces chloride and bicarbonate transport to epithelial lumen We believe Prime Editing-based approaches could eventually benefit more than 93% of all people with CF CFTR = cystic fibrosis transmembrane conductance regulator 23
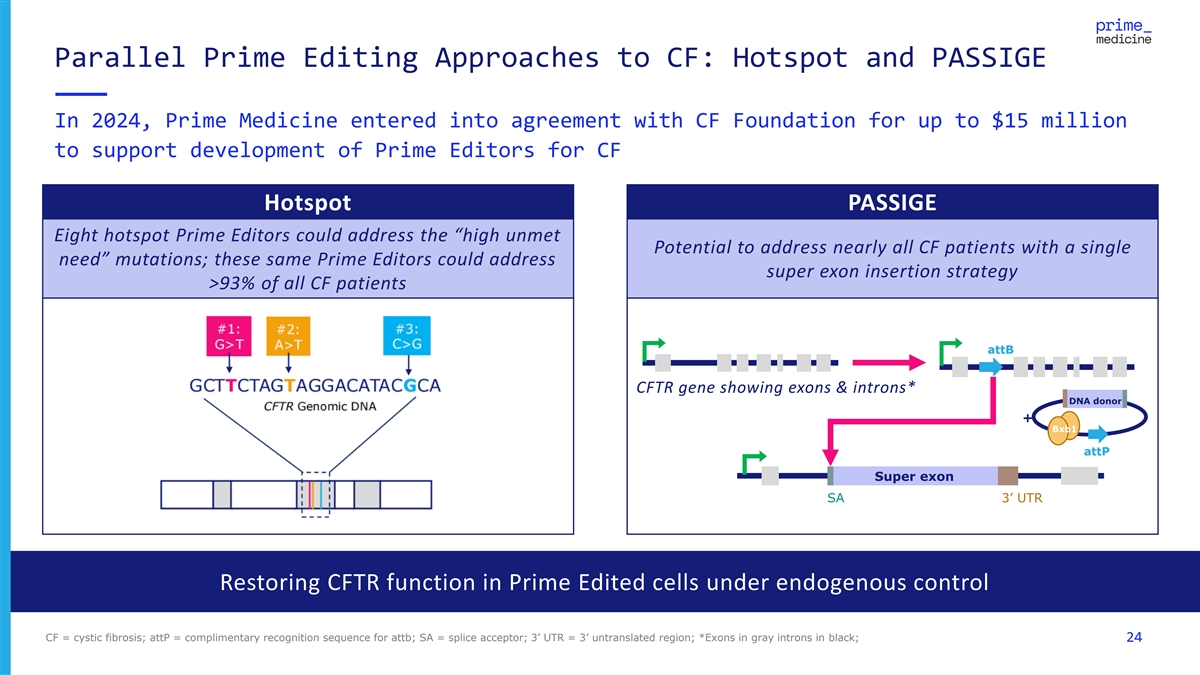
Parallel Prime Editing Approaches to CF: Hotspot and PASSIGE In 2024, Prime Medicine entered into agreement with CF Foundation for up to $15 million to support development of Prime Editors for CF Hotspot PASSIGE Eight hotspot Prime Editors could address the “high unmet Potential to address nearly all CF patients with a single need” mutations; these same Prime Editors could address super exon insertion strategy >93% of all CF patients attB CFTR gene showing exons & introns* DNA donor + Bxb1 attP Super exon SA 3’ UTR Restoring CFTR function in Prime Edited cells under endogenous control CF = cystic fibrosis; attP = complimentary recognition sequence for attb; SA = splice acceptor; 3’ UTR = 3’ untranslated region; *Exons in gray introns in black; 24
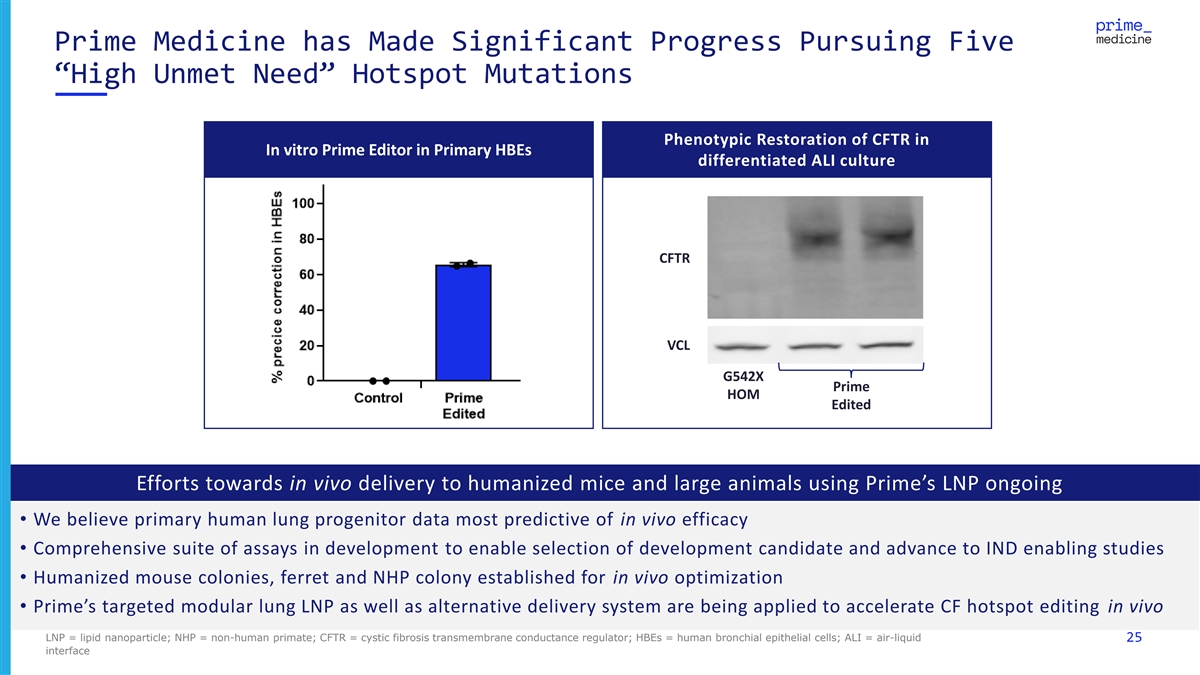
Prime Medicine has Made Significant Progress Pursuing Five “High Unmet Need” Hotspot Mutations Phenotypic Restoration of CFTR in In vitro Prime Editor in Primary HBEs differentiated ALI culture CFTR VCL G542X Prime HOM Edited Efforts towards in vivo delivery to humanized mice and large animals using Prime’s LNP ongoing • We believe primary human lung progenitor data most predictive of in vivo efficacy • Comprehensive suite of assays in development to enable selection of development candidate and advance to IND enabling studies • Humanized mouse colonies, ferret and NHP colony established for in vivo optimization • Prime’s targeted modular lung LNP as well as alternative delivery system are being applied to accelerate CF hotspot editing in vivo LNP = lipid nanoparticle; NHP = non-human primate; CFTR = cystic fibrosis transmembrane conductance regulator; HBEs = human bronchial epithelial cells; ALI = air-liquid 25 interface

Immunology and Oncology | 26 26

Strategic License and Broad Collaboration Agreement with Bristol Myers Squibb (BMS) to Develop Prime Edited ex Vivo CAR-T Products First broad, multi-target collaboration advancing Prime Editing for the treatment of complex oncology and autoimmune indications • $110 million upfront • >$3.5 billion in potential milestones, including: Leadership in Prime Editing; PASSIGE ‒ $185 million in preclinical milestones technology may enable one-step, non- viral, multi-kilobase-size editing ‒ $1.2 billion in development milestones approach with no double-stranded breaks ‒ More than $2.1 billion in commercial milestones Global leader in cell therapy for ‒ Royalties on net sales hematology, immunology and oncology • Multiple targets in immunological diseases and cancer, beyond the genetic diseases in Prime Medicine’s internal pipeline Prime Medicine retains rights to advance certain target reagents designed under this collaboration for applications beyond ex vivo T cell products, including in vivo T cell and other cell therapy applications 27
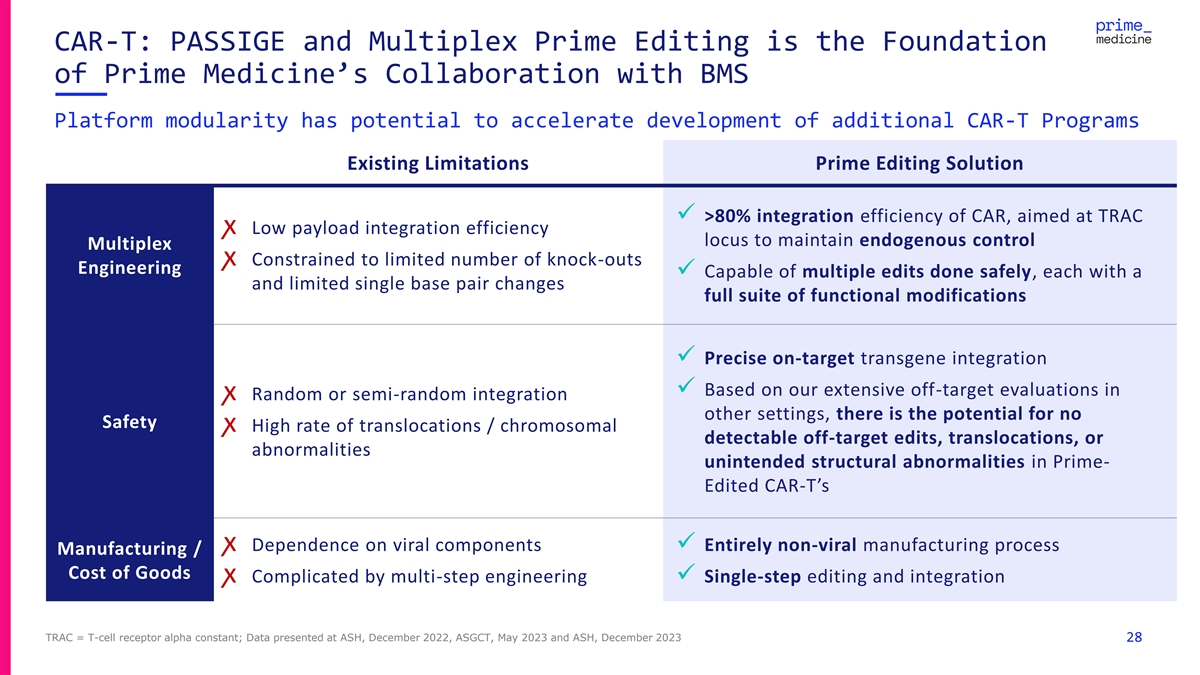
CAR-T: PASSIGE and Multiplex Prime Editing is the Foundation of Prime Medicine’s Collaboration with BMS Platform modularity has potential to accelerate development of additional CAR-T Programs Existing Limitations Prime Editing Solution ✓ >80% integration efficiency of CAR, aimed at TRAC ꭗ Low payload integration efficiency locus to maintain endogenous control Multiplex ꭗ Constrained to limited number of knock-outs Engineering ✓ Capable of multiple edits done safely, each with a and limited single base pair changes full suite of functional modifications ✓ Precise on-target transgene integration ✓ Based on our extensive off-target evaluations in ꭗ Random or semi-random integration other settings, there is the potential for no Safety ꭗ High rate of translocations / chromosomal detectable off-target edits, translocations, or abnormalities unintended structural abnormalities in Prime- Edited CAR-T’s ꭗ Dependence on viral components✓ Entirely non-viral manufacturing process Manufacturing / Cost of Goods ꭗ Complicated by multi-step engineering✓ Single-step editing and integration TRAC = T-cell receptor alpha constant; Data presented at ASH, December 2022, ASGCT, May 2023 and ASH, December 2023 28
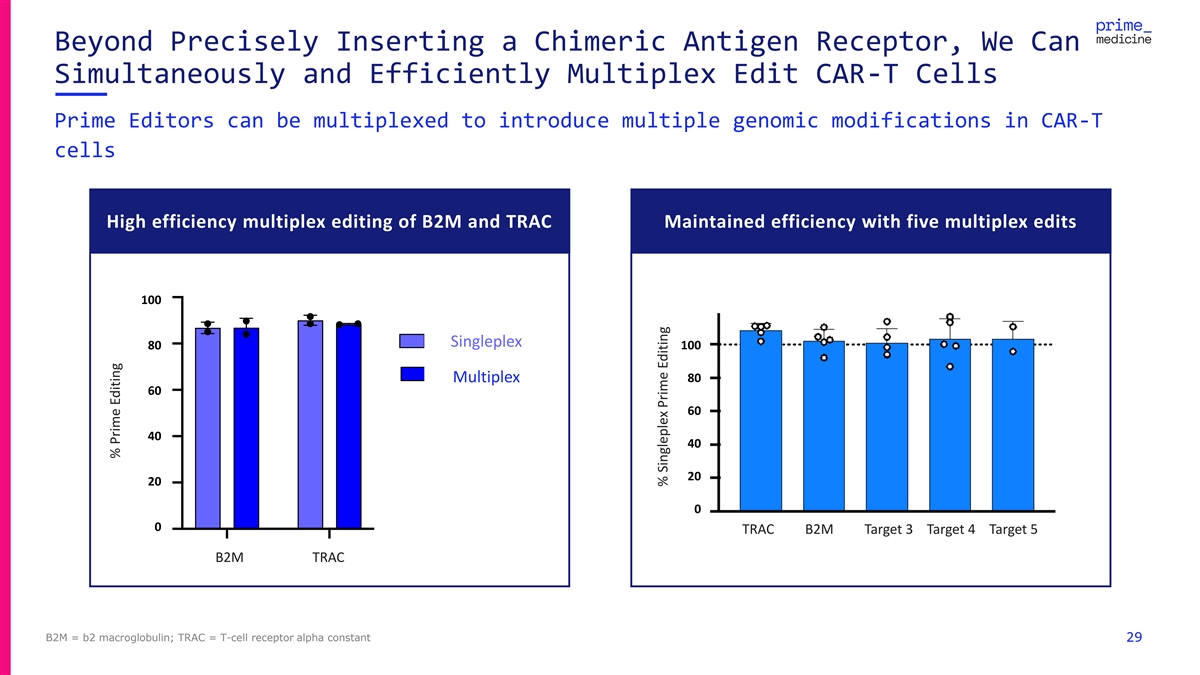
Beyond Precisely Inserting a Chimeric Antigen Receptor, We Can Simultaneously and Efficiently Multiplex Edit CAR-T Cells Prime Editors can be multiplexed to introduce multiple genomic modifications in CAR-T cells High efficiency multiplex editing of B2M and TRAC Maintained efficiency with five multiplex edits 100 100 SSi in ng glep leple le xx 80 80 100 Multiplex Multiplex 80 60 60 60 40 40 40 20 20 20 0 0 TRAC B2M Target 3 Target 4 Target 5 0 B2M TRAC B2M TRAC B2M = b2 macroglobulin; TRAC = T-cell receptor alpha constant 29 % Prime Editing % Prime Editing % Prime Editing % Singleplex Prime Editing

Corporate | 30 30
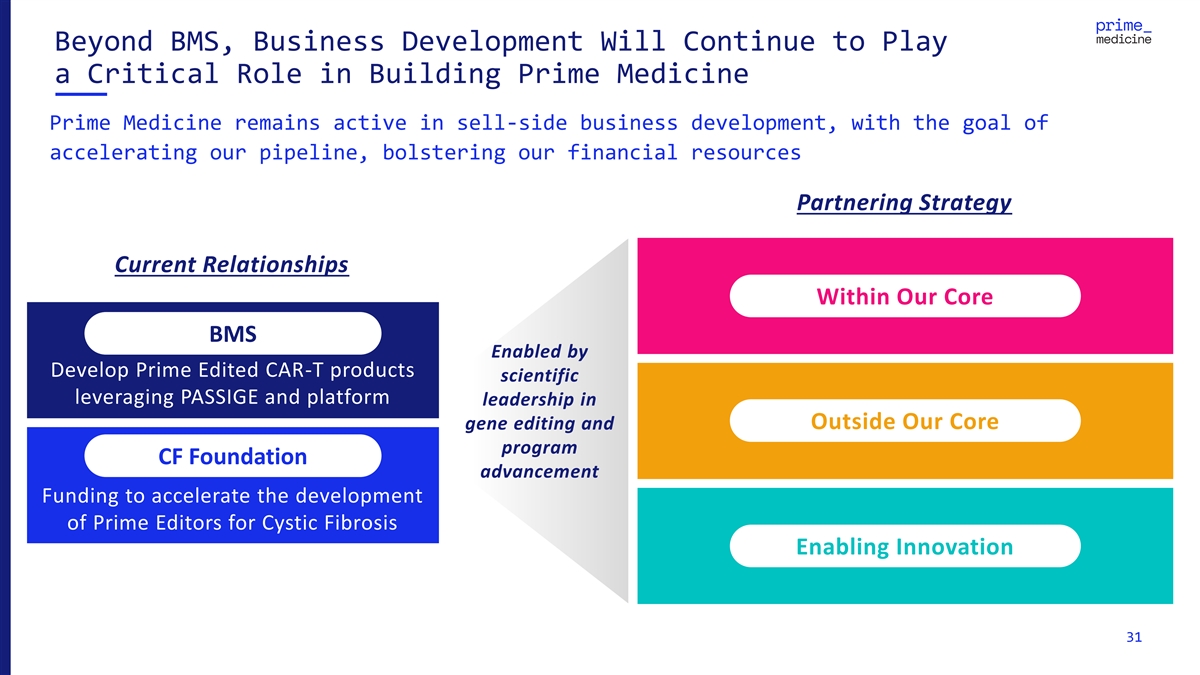
Beyond BMS, Business Development Will Continue to Play a Critical Role in Building Prime Medicine Prime Medicine remains active in sell-side business development, with the goal of accelerating our pipeline, bolstering our financial resources Partnering Strategy Current Relationships Within Our Core Within the Core BMS Enabled by Develop Prime Edited CAR-T products scientific leveraging PASSIGE and platform leadership in Outside Our Core gene editing and program CF Foundation advancement Funding to accelerate the development of Prime Editors for Cystic Fibrosis Enabling Innovation 31
Prime Medicine Holds Extensive, Foundational Intellectual Property for Prime Editing Technologies • Multiple configurations of RNA-templated gene editing - Prime Editor protein configurations: fusion, separate and split configurations - pegRNA configurations: fusion, split, separate and engineered configurations - Dual flap and dual guide RNA editing systems • Broad diversity of RNA-templated gene editing systems Seminal Prime Dual Flap Prime Editing PASSIGE System Editing Publication - Large variety of nucleic acid programmable DNA binding proteins - Extensive range of RNA-dependent DNA polymerases (reverse transcriptases) • PASSIGE: System using Prime Editing and recombinase to insert gene-sized DNA at chosen target location in genome - PASSIGE systems include various gene editing configurations and recombinases • Additional gene editing technology including DNA-dependent DNA PE4, PE5, PEmax Engineered pegRNAs pegRNA Enhancements polymerase editing • Program-specific patent filings for pipeline programs Prime Medicine holds 6 U.S. and 10 ex-U.S. issued patents - Numerous Prime-owned and in-licensed patent applications with broad coverage Improved Novel RT Proteins Novel PE Proteins recombinases filed worldwide - Pursuing aggressive filing strategy to cover technological advances pegRNA = Prime Editing guide RNA 32

Prime Medicine is the Leader in Gene Editing Positioned to Create Sustainable Value Through Pipeline Execution and External Partnerships 4 Potential to address more than 90% of genetic diseases and opportunities in non -genetic diseases The Leader in Prime 4 Pre-clinical efficacy across variety of target tissues leveraging various types of Prime Editing Editing 4 Comprehensive intellectual property position Platform 4 Fully integrated modular platform - pre-clinical, clinical, manufacturing, regulatory Modularity 4 Proprietary modular delivery systems within target tissues Oriented for 4 Advancing Prime Editing regulatory paradigms - streamlined development Growth Pipeline 4 PM577 in Wilson’s Disease IND and/or CTA expected in H1’26; AATD IND and/or CTA expected in mid-2026 Positioned 4 Strategically focused on programs in large genetic diseases, with clear path to value inflection and multi for Value billion-dollar opportunities Creation 4 BMS partnership to develop Prime Edited ex vivo CAR-T products Partnerships and BD 4 Cystic Fibrosis Foundation relationship and funding to advance Prime Editors for Cystic Fibrosis Potential 4 Additional business development to accelerate and expand pipeline Cash equivalents, investments and restricted cash of $158.3M as of 3/31/2025, cash runway into H1’26 AATD = Alpha-1 Antitrypsin Deficiency; IND = investigational new drug; CTA = clinical trial application 33

Appendix | 34 34
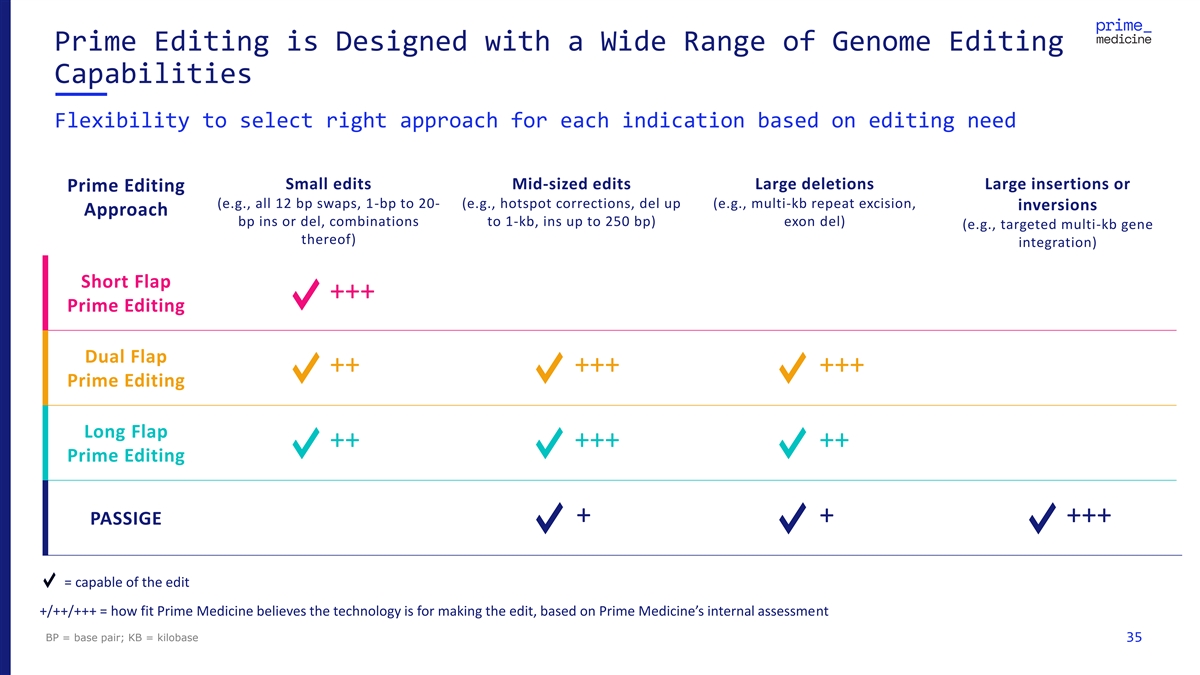
Prime Editing is Designed with a Wide Range of Genome Editing Capabilities Flexibility to select right approach for each indication based on editing need Small edits Mid-sized edits Large deletions Large insertions or Prime Editing (e.g., all 12 bp swaps, 1-bp to 20- (e.g., hotspot corrections, del up (e.g., multi-kb repeat excision, inversions Approach bp ins or del, combinations to 1-kb, ins up to 250 bp) exon del) (e.g., targeted multi-kb gene thereof) integration) Short Flap +++ Prime Editing Dual Flap ++ +++ +++ Prime Editing Long Flap ++ +++ ++ Prime Editing + + +++ PASSIGE = capable of the edit +/++/+++ = how fit Prime Medicine believes the technology is for making the edit, based on Prime Medicine’s internal assessment BP = base pair; KB = kilobase 35
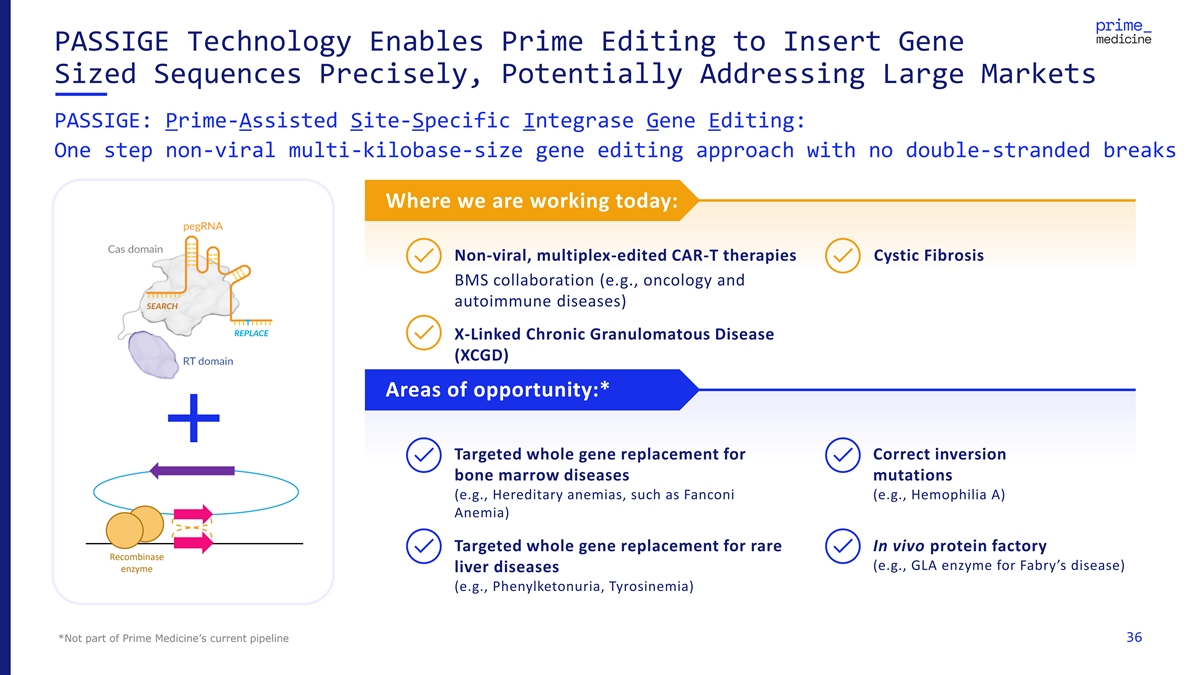
PASSIGE Technology Enables Prime Editing to Insert Gene Sized Sequences Precisely, Potentially Addressing Large Markets PASSIGE: Prime-Assisted Site-Specific Integrase Gene Editing: One step non-viral multi-kilobase-size gene editing approach with no double-stranded breaks Where we are working today: Non-viral, multiplex-edited CAR-T therapies Cystic Fibrosis BMS collaboration (e.g., oncology and autoimmune diseases) X-Linked Chronic Granulomatous Disease (XCGD) Areas of opportunity:* Targeted whole gene replacement for Correct inversion bone marrow diseases mutations (e.g., Hereditary anemias, such as Fanconi (e.g., Hemophilia A) Anemia) Targeted whole gene replacement for rare In vivo protein factory Recombinase (e.g., GLA enzyme for Fabry’s disease) enzyme liver diseases (e.g., Phenylketonuria, Tyrosinemia) *Not part of Prime Medicine’s current pipeline 36